Tumor‑associated macrophages activated in the tumor environment of hepatocellular carcinoma: Characterization and treatment (Review)
- PMID: 39239752
- PMCID: PMC11387121
- DOI: 10.3892/ijo.2024.5688
Tumor‑associated macrophages activated in the tumor environment of hepatocellular carcinoma: Characterization and treatment (Review)
Abstract
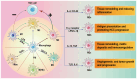
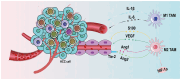
Hepatocellular carcinoma (HCC) tissue is rich in dendritic cells, T cells, B cells, macrophages, natural killer cells and cellular stroma. Together they form the tumor microenvironment (TME), which is also rich in numerous cytokines. Tumor‑associated macrophages (TAMs) are involved in the regulation of tumor development. TAMs in HCC receive stimuli in different directions, polarize in different directions and release different cytokines to regulate the development of HCC. TAMs are mostly divided into two cell phenotypes: M1 and M2. M1 TAMs secrete pro‑inflammatory mediators, and M2 TAMs secrete a variety of anti‑inflammatory and pro‑tumorigenic substances. The TAM polarization in HCC tumors is M2. Both direct and indirect methods for TAMs to regulate the development of HCC are discussed. TAMs indirectly support HCC development by promoting peripheral angiogenesis and regulating the immune microenvironment of the TME. In terms of the direct regulation between TAMs and HCC cells, the present review mainly focuses on the molecular mechanism. TAMs are involved in both the proliferation and apoptosis of HCC cells to regulate the quantitative changes of HCC, and stimulate the related invasive migratory ability and cell stemness of HCC cells. The present review aims to identify immunotherapeutic options based on the mechanisms of TAMs in the TME of HCC.
Keywords: hepatocellular carcinoma; immunotherapy; tumor microenvironment; tumorigenesis; tumor‑associated macrophages.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
