A monoclonal antibody targeting the Nipah virus fusion glycoprotein apex imparts protection from disease
- PMID: 39240113
- PMCID: PMC11494970
- DOI: 10.1128/jvi.00638-24
A monoclonal antibody targeting the Nipah virus fusion glycoprotein apex imparts protection from disease
Abstract
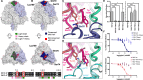
Nipah virus (NiV) is a highly pathogenic paramyxovirus capable of causing severe respiratory and neurologic disease in humans. Currently, there are no licensed vaccines or therapeutics against NiV, underscoring the urgent need for the development of countermeasures. The NiV surface-displayed glycoproteins, NiV-G and NiV-F, mediate host cell attachment and fusion, respectively, and are heavily targeted by host antibodies. Here, we describe a vaccination-derived neutralizing monoclonal antibody, mAb92, that targets NiV-F. Structural characterization of the Fab region bound to NiV-F (NiV-F-Fab92) by cryo-electron microscopy analysis reveals an epitope in the DIII domain at the membrane distal apex of NiV-F, an established site of vulnerability on the NiV surface. Further, prophylactic treatment of hamsters with mAb92 offered complete protection from NiV disease, demonstrating beneficial activity of mAb92 in vivo. This work provides support for targeting NiV-F in the development of vaccines and therapeutics against NiV.IMPORTANCENipah virus (NiV) is a highly lethal henipavirus (HNV) that causes severe respiratory and neurologic disease in humans. Currently, there are no licensed vaccines or therapeutics against NiV, highlighting a need to develop countermeasures. The NiV surface displays the receptor binding protein (NiV-G, or RBP) and the fusion protein (NiV-F), which allow the virus to attach and enter cells. These proteins can be targeted by vaccines and antibodies to prevent disease. This work describes a neutralizing antibody (mAb92) that targets NiV-F. Structural characterization by cryo-electron microscopy analysis reveals where the antibody binds to NiV-F to neutralize the virus. This study also shows that prophylactic treatment of hamsters with mAb92 completely protected against developing NiV disease. This work shows how targeting NiV-F can be useful to preventing NiV disease, supporting future studies in the development of vaccines and therapeutics.
Keywords: Nipah virus; fusion glycoprotein; henipavirus; monoclonal antibodies; neutralizing antibodies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






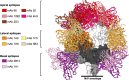
References
-
- Hossain MJ, Gurley ES, Montgomery JM, Bell M, Carroll DS, Hsu VP, Formenty P, Croisier A, Bertherat E, Faiz MA, Azad AK, Islam R, Molla MAR, Ksiazek TG, Rota PA, Comer JA, Rollin PE, Luby SP, Breiman RF. 2008. Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis 46:977–984. doi: 10.1086/529147 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

