Exposome-Wide Ranking to Uncover Environmental Chemicals Associated with Dyslipidemia: A Panel Study in Healthy Older Chinese Adults from the BAPE Study
- PMID: 39240788
- PMCID: PMC11379127
- DOI: 10.1289/EHP13864
Exposome-Wide Ranking to Uncover Environmental Chemicals Associated with Dyslipidemia: A Panel Study in Healthy Older Chinese Adults from the BAPE Study
Abstract
Background: Environmental contaminants (ECs) are increasingly recognized as crucial drivers of dyslipidemia and cardiovascular disease (CVD), but the comprehensive impact spectrum and interlinking mechanisms remain uncertain.
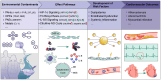
Objectives: We aimed to systematically evaluate the association between exposure to 80 ECs across seven divergent categories and markers of dyslipidemia and investigate their underpinning biomolecular mechanisms via an unbiased integrative approach of internal chemical exposome and multi-omics.
Methods: A longitudinal study involving 76 healthy older adults was conducted in Jinan, China, and participants were followed five times from 10 September 2018 to 19 January 2019 in 1-month intervals. A broad spectrum of seven chemical categories covering the prototypes and metabolites of 102 ECs in serum or urine as well as six serum dyslipidemia markers [total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, apolipoprotein (Apo)A1, ApoB, and ApoE4] were measured. Multi-omics, including the blood transcriptome, serum/urine metabolome, and serum lipidome, were profiled concurrently. Exposome-wide association study and the deletion/substitution/addition algorithms were applied to explore the associations between 80 EC exposures detection frequency and dyslipidemia markers. Weighted quantile sum regression was used to assess the mixture effects and relative contributions. Multi-omics profiling, causal inference model, and pathway analysis were conducted to interpret the mediating biomolecules and underlying mechanisms. Examination of cytokines and electrocardiograms was further conducted to validate the observed associations and biomolecular pathways.
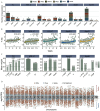
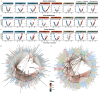
Results: Eight main ECs [1-naphthalene, 1-pyrene, 2-fluorene, dibutyl phosphate, tri-phenyl phosphate, mono-(2-ethyl-5-hydroxyhexyl) phthalate, chromium, and vanadium] were significantly associated with most dyslipidemia markers. Multi-omics indicated that the associations were mediated by endogenous biomolecules and pathways, primarily pertinent to CVD, inflammation, and metabolism. Clinical measures of cytokines and electrocardiograms further cross-validated the association of these exogenous ECs with systemic inflammation and cardiac function, demonstrating their potential mechanisms in driving dyslipidemia pathogenesis.
Discussion: It is imperative to prioritize mitigating exposure to these ECs in the primary prevention and control of the dyslipidemia epidemic. https://doi.org/10.1289/EHP13864.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
