The triglyceride-synthesizing enzyme diacylglycerol acyltransferase 2 modulates the formation of the hepatitis C virus replication organelle
- PMID: 39241103
- PMCID: PMC11410266
- DOI: 10.1371/journal.ppat.1012509
The triglyceride-synthesizing enzyme diacylglycerol acyltransferase 2 modulates the formation of the hepatitis C virus replication organelle
Abstract
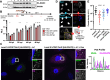
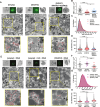
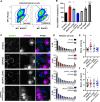
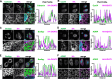
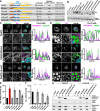
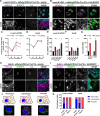
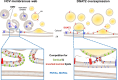
The replication organelle of hepatitis C virus (HCV), called membranous web, is derived from the endoplasmic reticulum (ER) and mainly comprises double membrane vesicles (DMVs) that concentrate the viral replication complexes. It also tightly associates with lipid droplets (LDs), which are essential for virion morphogenesis. In particular acyl-CoA:diacylglycerol acyltransferase 1 (DGAT1), a rate-limiting enzyme in triglyceride synthesis, promotes early steps of virus assembly. The close proximity between ER membranes, DMVs and LDs therefore permits the efficient coordination of the HCV replication cycle. Here, we demonstrate that exaggerated LD accumulation due to the excessive expression of the DGAT1 isozyme, DGAT2, dramatically impairs the formation of the HCV membranous web. This effect depended on the enzymatic activity and ER association of DGAT2, whereas the mere LD accumulation was not sufficient to hamper HCV RNA replication. Our lipidomics data indicate that both HCV infection and DGAT2 overexpression induced membrane lipid biogenesis and markedly increased phospholipids with long chain polyunsaturated fatty acids, suggesting a dual use of these lipids and their possible competition for LD and DMV biogenesis. On the other hand, overexpression of DGAT2 depleted specific phospholipids, particularly oleyl fatty acyl chain-containing phosphatidylcholines, which, in contrast, are increased in HCV-infected cells and likely essential for viral infection. In conclusion, our results indicate that lipid exchanges occurring during LD biogenesis regulate the composition of intracellular membranes and thereby affect the formation of the HCV replication organelle. The potent antiviral effect observed in our DGAT2 overexpression system unveils lipid flux that may be relevant in the context of steatohepatitis, a hallmark of HCV infection, but also in physiological conditions, locally in specific subdomains of the ER membrane. Thus, LD formation mediated by DGAT1 and DGAT2 might participate in the spatial compartmentalization of HCV replication and assembly factories within the membranous web.
Copyright: © 2024 Reichert et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

