Human Smc5/6 recognises transcription-generated positive DNA supercoils
- PMID: 39242537
- PMCID: PMC11379904
- DOI: 10.1038/s41467-024-50646-w
Human Smc5/6 recognises transcription-generated positive DNA supercoils
Abstract
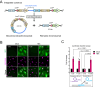
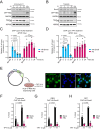
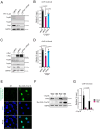
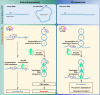
Beyond its essential roles in ensuring faithful chromosome segregation and genomic stability, the human Smc5/6 complex acts as an antiviral factor. It binds to and impedes the transcription of extrachromosomal DNA templates; an ability which is lost upon integration of the DNA into the chromosome. How the complex distinguishes among different DNA templates is unknown. Here we show that, in human cells, Smc5/6 preferentially binds to circular rather than linear extrachromosomal DNA. We further demonstrate that the transcriptional process, per se, and particularly the accumulation of DNA secondary structures known to be substrates for topoisomerases, is responsible for Smc5/6 recruitment. More specifically, we find that in vivo Smc5/6 binds to positively supercoiled DNA. Those findings, in conjunction with our genome-wide Smc5/6 binding analysis showing that Smc5/6 localizes at few but highly transcribed chromosome loci, not only unveil a previously unforeseen role of Smc5/6 in DNA topology management during transcription but highlight the significance of sensing DNA topology as an antiviral defense mechanism.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

