Glutathione peroxidase 3 is a potential biomarker for konzo
- PMID: 39242582
- PMCID: PMC11379914
- DOI: 10.1038/s41467-024-52136-5
Glutathione peroxidase 3 is a potential biomarker for konzo
Abstract
Konzo is a neglected paralytic neurological disease associated with food (cassava) poisoning that affects the world's poorest children and women of childbearing ages across regions of sub-Saharan Africa. Despite understanding the dietary factors that lead to konzo, the molecular markers and mechanisms that trigger this disease remain unknown. To identify potential protein biomarkers associated with a disease status, plasma was collected from two independent Congolese cohorts, a discovery cohort (n = 60) and validation cohort (n = 204), sampled 10 years apart and subjected to multiple high-throughput assays. We identified that Glutathione Peroxidase 3 (GPx3), a critical plasma-based antioxidant enzyme, was the sole protein examined that was both significantly and differentially abundant between affected and non-affected participants in both cohorts, with large reductions observed in those affected with konzo. Our findings raise the notion that reductions in key antioxidant mechanisms may be the biological risk factor for the development of konzo, particularly those mediated through pathways involving the glutathione peroxidase family.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
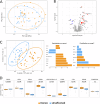
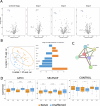
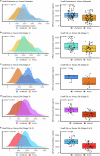
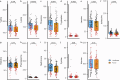
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

