Hyper-recombination in ribosomal DNA is driven by long-range resection-independent RAD51 accumulation
- PMID: 39242676
- PMCID: PMC11379943
- DOI: 10.1038/s41467-024-52189-6
Hyper-recombination in ribosomal DNA is driven by long-range resection-independent RAD51 accumulation
Abstract
Ribosomal DNA (rDNA) encodes the ribosomal RNA genes and represents an intrinsically unstable genomic region. However, the underlying mechanisms and implications for genome integrity remain elusive. Here, we use Bloom syndrome (BS), a rare genetic disease characterized by DNA repair defects and hyper-unstable rDNA, as a model to investigate the mechanisms leading to rDNA instability. We find that in Bloom helicase (BLM) proficient cells, the homologous recombination (HR) pathway in rDNA resembles that in nuclear chromatin; it is initiated by resection, replication protein A (RPA) loading and BRCA2-dependent RAD51 filament formation. However, BLM deficiency compromises RPA-loading and BRCA1/2 recruitment to rDNA, but not RAD51 accumulation. RAD51 accumulates at rDNA despite depletion of long-range resection nucleases and rDNA damage results in micronuclei when BLM is absent. In summary, our findings indicate that rDNA is permissive to RAD51 accumulation in the absence of BLM, leading to micronucleation and potentially global genomic instability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
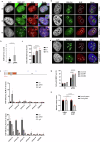

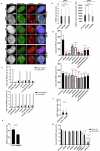
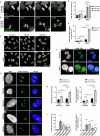
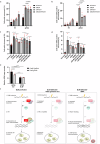
References
Publication types
MeSH terms
Substances
Grants and funding
- R311-A18224/Kræftens Bekæmpelse (Danish Cancer Society)
- A10965/Kræftens Bekæmpelse (Danish Cancer Society)
- R167-A11068/Kræftens Bekæmpelse (Danish Cancer Society)
- R311-A18224/Kræftens Bekæmpelse (Danish Cancer Society)
- NNF18OC0052647/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF22OC0078971/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF21OC0068988, NNF21OC0072031/Novo Nordisk Fonden (Novo Nordisk Foundation)
- 8045-00057A/Det Frie Forskningsråd (Danish Council for Independent Research)
- 8045-00057A/Det Frie Forskningsråd (Danish Council for Independent Research)
- 8048-00072A/Det Frie Forskningsråd (Danish Council for Independent Research)
- 1026-00241B/Det Frie Forskningsråd (Danish Council for Independent Research)
- R347-2020-2219/Lundbeckfonden (Lundbeck Foundation)
- R322-2019-2577/Lundbeckfonden (Lundbeck Foundation)
- VR-MH 2014-46602-117891-30/Vetenskapsrådet (Swedish Research Council)
- 899417/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

