Continental-scale associations of Arabidopsis thaliana phyllosphere members with host genotype and drought
- PMID: 39242816
- PMCID: PMC11457713
- DOI: 10.1038/s41564-024-01773-z
Continental-scale associations of Arabidopsis thaliana phyllosphere members with host genotype and drought
Abstract
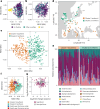
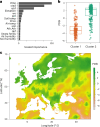
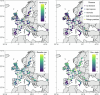
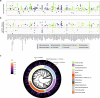
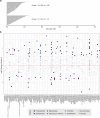
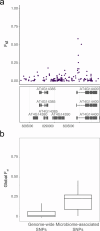
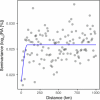
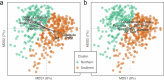
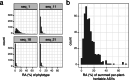
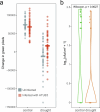
Plants are colonized by distinct pathogenic and commensal microbiomes across different regions of the globe, but the factors driving their geographic variation are largely unknown. Here, using 16S ribosomal DNA and shotgun sequencing, we characterized the associations of the Arabidopsis thaliana leaf microbiome with host genetics and climate variables from 267 populations in the species' native range across Europe. Comparing the distribution of the 575 major bacterial amplicon variants (phylotypes), we discovered that microbiome composition in A. thaliana segregates along a latitudinal gradient. The latitudinal clines in microbiome composition are predicted by metrics of drought, but also by the spatial genetics of the host. To validate the relative effects of drought and host genotype we conducted a common garden field study, finding 10% of the core bacteria to be affected directly by drought and 20% to be affected by host genetic associations with drought. These data provide a valuable resource for the plant microbiome field, with the identified associations suggesting that drought can directly and indirectly shape genetic variation in A. thaliana via the leaf microbiome.
© 2024. The Author(s).
Conflict of interest statement
D.W. holds equity in Computomics, which advises plant breeders. D.W. consults for KWS SE, a plant breeder and seed producer. The other authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- PATHOCOM/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- T32 GM141848/GM/NIGMS NIH HHS/United States
- LTF/Human Frontier Science Program (HFSP)
- R35 GM150722/GM/NIGMS NIH HHS/United States
- Startup funds/University of Utah (U of U)

