Loss of DOCK2 potentiates Inflammatory Bowel Disease-associated colorectal cancer via immune dysfunction and IFNγ induction of IDO1 expression
- PMID: 39242821
- PMCID: PMC11473400
- DOI: 10.1038/s41388-024-03135-9
Loss of DOCK2 potentiates Inflammatory Bowel Disease-associated colorectal cancer via immune dysfunction and IFNγ induction of IDO1 expression
Abstract
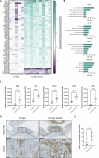
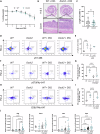
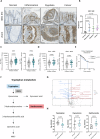
Inflammatory Bowel Disease-associated colorectal cancer (IBD-CRC) is a known and serious complication of Inflammatory Bowel Disease (IBD) affecting the colon. However, relatively little is known about the pathogenesis of IBD-associated colorectal cancer in comparison with its sporadic cancer counterpart. Here, we investigated the function of Dock2, a gene mutated in ~10% of IBD-associated colorectal cancers that encodes a guanine nucleotide exchange factor (GEF). Using a genetically engineered mouse model of IBD-CRC, we found that whole body loss of Dock2 increases tumourigenesis via immune dysregulation. Dock2-deficient tumours displayed increased levels of IFNγ-associated genes, including the tryptophan metabolising, immune modulatory enzyme, IDO1, when compared to Dock2-proficient tumours. This phenotype was driven by increased IFNγ-production in T cell populations, which infiltrated Dock2-deficient tumours, promoting IDO1 expression in tumour epithelial cells. We show that IDO1 inhibition delays tumourigenesis in Dock2 knockout mice, and we confirm that this pathway is conserved across species as IDO1 expression is elevated in human IBD-CRC and in sporadic CRC cases with mutated DOCK2. Together, these data demonstrate a previously unidentified tumour suppressive role of DOCK2 that limits IFNγ-induced IDO1 expression and cancer progression, opening potential new avenues for therapeutic intervention.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease. N Engl J Med. 2015;372:1441–52. - PubMed
-
- Coskun M, Vermeire S, Nielsen OH. Novel targeted therapies for inflammatory bowel disease. Trends Pharmacol Sci. 2017;38:127–42. - PubMed
-
- Jewel Samadder N, Valentine JF, Guthery S, Singh H, Bernstein CN, Wan Y, et al. Colorectal cancer in inflammatory bowel diseases: a population-based study in Utah. Dig Dis Sci. 2017;62:2126–32. - PubMed
-
- Olen O, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, et al. Colorectal cancer in ulcerative colitis: a Scandinavian population-based cohort study. Lancet. 2020;395:123–31. - PubMed
MeSH terms
Substances
Grants and funding
- GDCOLCA 800112/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 214369/Z/18/Z/Wellcome Trust (Wellcome)
- 715782/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- A19166/Cancer Research UK (CRUK)
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

