A systematic review and meta-analysis on the transcriptomic signatures in alcohol use disorder
- PMID: 39242950
- PMCID: PMC11649567
- DOI: 10.1038/s41380-024-02719-x
A systematic review and meta-analysis on the transcriptomic signatures in alcohol use disorder
Abstract
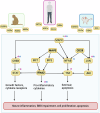
Currently available clinical treatments on alcohol use disorder (AUD) exhibit limited efficacy and new druggable targets are required. One promising approach to discover new molecular treatment targets involves the transcriptomic profiling of brain regions within the addiction neurocircuitry, utilizing animal models and postmortem brain tissue from deceased patients with AUD. Unfortunately, such studies suffer from large heterogeneity and small sample sizes. To address these limitations, we conducted a cross-species meta-analysis on transcriptome-wide data obtained from brain tissue of patients with AUD and animal models. We integrated 36 cross-species transcriptome-wide RNA-expression datasets with an alcohol-dependent phenotype vs. controls, following the PRISMA guidelines. In total, we meta-analyzed 964 samples - 502 samples from the prefrontal cortex (PFC), 282 nucleus accumbens (NAc) samples, and 180 from amygdala (AMY). The PFC had the highest number of differentially expressed genes (DEGs) across rodents, monkeys, and humans. Commonly dysregulated DEGs suggest conserved cross-species mechanisms for chronic alcohol consumption/AUD comprising MAPKs as well as STAT, IRF7, and TNF. Furthermore, we identified numerous unique gene sets that might contribute individually to these conserved mechanisms and also suggest novel molecular aspects of AUD. Validation of the transcriptomic alterations on the protein level revealed interesting targets for further investigation. Finally, we identified a combination of DEGs that are commonly regulated across different brain tissues as potential biomarkers for AUD. In summary, we provide a compendium of genes that are assessable via a shiny app, and describe signaling pathways, and physiological and cellular processes that are altered in AUD that require future studies for functional validation.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Alcohol: Fact sheet. https://www.who.int/en/news-room/fact-sheets/detail/alcohol, 2018.
Publication types
MeSH terms
Grants and funding
- P51 OD011092/OD/NIH HHS/United States
- TRR265/Deutsche Forschungsgemeinschaft (German Research Foundation)
- P60AA010760/U.S. Department of Health & Human Services | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- P60 AA010760/AA/NIAAA NIH HHS/United States
- OD P51OD011092/U.S. Department of Health & Human Services | NIH | NIH Clinical Center (Clinical Center)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

