RNA splicing factor RBFOX2 is a key factor in the progression of cancer and cardiomyopathy
- PMID: 39243148
- PMCID: PMC11380049
- DOI: 10.1002/ctm2.1788
RNA splicing factor RBFOX2 is a key factor in the progression of cancer and cardiomyopathy
Abstract
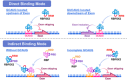
Background: Alternative splicing of pre-mRNA is a fundamental regulatory process in multicellular eukaryotes, significantly contributing to the diversification of the human proteome. RNA-binding fox-1 homologue 2 (RBFOX2), a member of the evolutionarily conserved RBFOX family, has emerged as a critical splicing regulator, playing a pivotal role in the alternative splicing of pre-mRNA. This review provides a comprehensive analysis of RBFOX2, elucidating its splicing activity through direct and indirect binding mechanisms. RBFOX2 exerts substantial influence over the alternative splicing of numerous transcripts, thereby shaping essential cellular processes such as differentiation and development.
Main body of the abstract: Dysregulation of RBFOX2-mediated alternative splicing has been closely linked to a spectrum of cardiovascular diseases and malignant tumours, underscoring its potential as a therapeutic target. Despite significant progress, current research faces notable challenges. The complete structural characterisation of RBFOX2 remains elusive, limiting in-depth exploration beyond its RNA-recognition motif. Furthermore, the scarcity of studies focusing on RBFOX2-targeting drugs poses a hindrance to translating research findings into clinical applications.
Conclusion: This review critically assesses the existing body of knowledge on RBFOX2, highlighting research gaps and limitations. By delineating these areas, this analysis not only serves as a foundational reference for future studies but also provides strategic insights for bridging these gaps. Addressing these challenges will be instrumental in unlocking the full therapeutic potential of RBFOX2, paving the way for innovative and effective treatments in various diseases.
Keywords: RBFOX2; alternative splicing; pre‐mRNA; therapeutic targeting.
© 2024 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures




Similar articles
-
RBFOX2 protein domains and cellular activities.Biochem Soc Trans. 2014 Aug;42(4):1180-3. doi: 10.1042/BST20140050. Biochem Soc Trans. 2014. PMID: 25110022 Review.
-
RBFOX2 alters splicing outcome in distinct binding modes with multiple protein partners.Nucleic Acids Res. 2021 Aug 20;49(14):8370-8383. doi: 10.1093/nar/gkab595. Nucleic Acids Res. 2021. PMID: 34244793 Free PMC article.
-
Genes of the cGMP-PKG-Ca2+ signaling pathway are alternatively spliced in cardiomyopathy: Role of RBFOX2.Biochim Biophys Acta Mol Basis Dis. 2020 Mar 1;1866(3):165620. doi: 10.1016/j.bbadis.2019.165620. Epub 2019 Nov 25. Biochim Biophys Acta Mol Basis Dis. 2020. PMID: 31778749 Free PMC article.
-
RBFOX2 deregulation promotes pancreatic cancer progression and metastasis through alternative splicing.Nat Commun. 2023 Dec 19;14(1):8444. doi: 10.1038/s41467-023-44126-w. Nat Commun. 2023. PMID: 38114498 Free PMC article.
-
RBFOX2 as a regulatory linchpin in cancer: insights from a comprehensive review of its roles in tumorigenesis.Am J Cancer Res. 2024 Oct 25;14(10):5045-5060. doi: 10.62347/BNPO2363. eCollection 2024. Am J Cancer Res. 2024. PMID: 39553227 Free PMC article. Review.
Cited by
-
LncRNA HSCHARME is altered in human cardiomyopathies and promotes stem cell-derived cardiomyogenesis via splicing regulation.Nat Commun. 2025 Aug 23;16(1):7880. doi: 10.1038/s41467-025-62754-2. Nat Commun. 2025. PMID: 40849301 Free PMC article.
-
Upregulation of WDR4 mediated by RBFOX2 promotes laryngeal cancer progression through the WDR4/m7G/lncRNA ZFAS1/RBFOX2 axis.Naunyn Schmiedebergs Arch Pharmacol. 2025 Jun;398(6):7529-7543. doi: 10.1007/s00210-024-03779-0. Epub 2025 Jan 7. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 39774908
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
