Multicenter radio-multiomic analysis for predicting breast cancer outcome and unravelling imaging-biological connection
- PMID: 39244594
- PMCID: PMC11380684
- DOI: 10.1038/s41698-024-00666-y
Multicenter radio-multiomic analysis for predicting breast cancer outcome and unravelling imaging-biological connection
Abstract
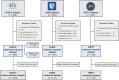
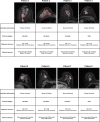
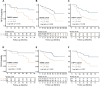
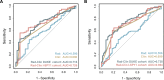
Radiomics offers a noninvasive avenue for predicting clinicopathological factors. However, thorough investigations into a robust breast cancer outcome-predicting model and its biological significance remain limited. This study develops a robust radiomic model for prognosis prediction, and further excavates its biological foundation and transferring prediction performance. We retrospectively collected preoperative dynamic contrast-enhanced MRI data from three distinct breast cancer patient cohorts. In FUSCC cohort (n = 466), Lasso was used to select features correlated with patient prognosis and multivariate Cox regression was utilized to integrate these features and build the radiomic risk model, while multiomic analysis was conducted to investigate the model's biological implications. DUKE cohort (n = 619) and I-SPY1 cohort (n = 128) were used to test the performance of the radiomic signature in outcome prediction. A thirteen-feature radiomic signature was identified in the FUSCC cohort training set and validated in the FUSCC cohort testing set, DUKE cohort and I-SPY1 cohort for predicting relapse-free survival (RFS) and overall survival (OS) (RFS: p = 0.013, p = 0.024 and p = 0.035; OS: p = 0.036, p = 0.005 and p = 0.027 in the three cohorts). Multiomic analysis uncovered metabolic dysregulation underlying the radiomic signature (ATP metabolic process: NES = 1.84, p-adjust = 0.02; cholesterol biosynthesis: NES = 1.79, p-adjust = 0.01). Regarding the therapeutic implications, the radiomic signature exhibited value when combining clinical factors for predicting the pathological complete response to neoadjuvant chemotherapy (DUKE cohort, AUC = 0.72; I-SPY1 cohort, AUC = 0.73). In conclusion, our study identified a breast cancer outcome-predicting radiomic signature in a multicenter radio-multiomic study, along with its correlations with multiomic features in prognostic risk assessment, laying the groundwork for future prospective clinical trials in personalized risk stratification and precision therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality Worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin.71, 209–249 (2021). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

