Genome-wide identification of cannabinoid biosynthesis genes in non-drug type Cannabis (Cannabis sativa L.) cultivar
- PMID: 39244597
- PMCID: PMC11380790
- DOI: 10.1186/s42238-024-00246-8
Genome-wide identification of cannabinoid biosynthesis genes in non-drug type Cannabis (Cannabis sativa L.) cultivar
Abstract
Background: Cannabis sativa cultivars can be classified as marijuana or hemp, depending on its amount of the psychoactive cannabinoid Δ9-tetrahydrocannabinolic acid (THCA). Hemp Cheungsam is a non-drug type Cannabis sativa that is characterized by low THCA content. However, the transcripts and expression profile of cannabinoid biosynthesis pathway genes of hemp Cheungsam have not been investigated.
Methods: RNA-sequencing (RNA-seq) was performed on three different tissue types (flower, leaf, and stem) of hemp Cheungsam to understand their transcriptomes. The expression of cannabinoid biosynthesis pathway genes was further analyzed in each tissue type. Multiple sequence alignment and conserved domain analyses were used to investigate the homologs of cannbinoid biosynthesis genes.
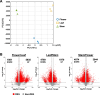
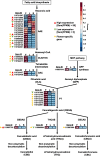
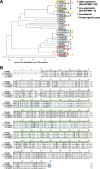
Results: We found that the cannabinoid biosynthesis pathway was mainly expressed in the flowers of hemp Cheungsam, similar to other Cannabis cultivars. However, expression of cannabidiolic acid (CBDA) synthase was much higher than THCA synthase and cannabichromenic acid (CBCA) synthase, suggesting that the transcription profile favors CBDA biosynthesis. Sequence analysis of cannabinoid biosynthesis pathway genes suggested the identity of orthologs in hemp Cheungsam.
Conclusions: Cannabinoid biosynthesis in hemp Cheungsam mostly occurs in the flowers, compared to other plant organs. While CBDA synthase expression is high, THCA and CBCA synthase expression is considerably low, indicating lesser THCA biosynthesis and thus low THCA content. Sequence analysis of key genes (CBDA, THCA, and CBCA synthases) of the cannabinoid biosynthetic pathway indicates that orthologs are present in hemp Cheungsam.
Keywords: Cannabis sativa; Hemp; RNA-seq.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Evaluation of tetrahydrocannabinolic acid (THCA) synthase polymorphisms for distinguishing between marijuana and hemp.J Forensic Sci. 2022 Jul;67(4):1370-1381. doi: 10.1111/1556-4029.15045. Epub 2022 Apr 13. J Forensic Sci. 2022. PMID: 35416290
-
The draft genome and transcriptome of Cannabis sativa.Genome Biol. 2011 Oct 20;12(10):R102. doi: 10.1186/gb-2011-12-10-r102. Genome Biol. 2011. PMID: 22014239 Free PMC article.
-
Elucidation of structure-function relationship of THCA and CBDA synthase from Cannabis sativaL.J Biotechnol. 2018 Oct 20;284:17-26. doi: 10.1016/j.jbiotec.2018.07.031. Epub 2018 Jul 24. J Biotechnol. 2018. PMID: 30053500
-
Comprehending and improving cannabis specialized metabolism in the systems biology era.Plant Sci. 2020 Sep;298:110571. doi: 10.1016/j.plantsci.2020.110571. Epub 2020 Jun 27. Plant Sci. 2020. PMID: 32771172 Review.
-
Recent advances in Cannabis sativa research: biosynthetic studies and its potential in biotechnology.Curr Pharm Biotechnol. 2007 Aug;8(4):237-43. doi: 10.2174/138920107781387456. Curr Pharm Biotechnol. 2007. PMID: 17691992 Review.
References
-
- Aloo SO, Kwame FO, Oh D-H. Identification of possible bioactive compounds and a comparative study on in vitro biological properties of whole hemp seed and stem. Food Biosci. 2023b;51: 102329.10.1016/j.fbio.2022.102329 - DOI
-
- Anderson PJ, de la Paz A. How to identify hemp, Cannabis sativa L. (and lookalike) plants. Florida Department of Agriculture and Consumer Services Circular 43. 2021. https://www.fdacs.gov/ezs3download/download/96962/2651285/Media/Files/Pl.... Accessed 25 May 2023.
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 14 Mar 2022.
LinkOut - more resources
Full Text Sources

