Development of an automated 3D high content cell screening platform for organoid phenotyping
- PMID: 39245180
- PMCID: PMC12380041
- DOI: 10.1016/j.slasd.2024.100182
Development of an automated 3D high content cell screening platform for organoid phenotyping
Abstract
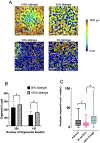
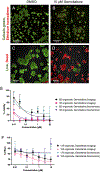
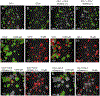
The use of organoid models in biomedical research has grown substantially since their inception. As they gain popularity among scientists seeking more complex and biologically relevant systems, there is a direct need to expand and clarify potential uses of such systems in diverse experimental contexts. Herein we outline a high-content screening (HCS) platform that allows researchers to screen drugs or other compounds against three-dimensional (3D) cell culture systems in a multi-well format (384-well). Furthermore, we compare the quality of robotic liquid handling with manual pipetting and characterize and contrast the phenotypic effects detected by confocal imaging and biochemical assays in response to drug treatment. We show that robotic liquid handling is more consistent and amendable to high throughput experimental designs when compared to manual pipetting due to improved precision and automated randomization capabilities. We also show that image-based techniques are more sensitive to detecting phenotypic changes within organoid cultures than traditional biochemical assays that evaluate cell viability, supporting their integration into organoid screening workflows. Finally, we highlight the enhanced capabilities of confocal imaging in this organoid screening platform as they relate to discerning organoid drug responses in single-well co-cultures of organoids derived from primary human biopsies and patient-derived xenograft (PDX) models. Altogether, this platform enables automated, imaging-based HCS of 3D cellular models in a non-destructive manner, opening the path to complementary analysis through integrated downstream methods.
Keywords: 3D tissue culture; Automation; High-content imaging; High-content screening; Image-based phenotyping; Organoid co-culture.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

