Discovery and optimisation of pyrazolo[1,5- a]pyrimidines as aryl hydrocarbon receptor antagonists
- PMID: 39246744
- PMCID: PMC11376203
- DOI: 10.1039/d4md00266k
Discovery and optimisation of pyrazolo[1,5- a]pyrimidines as aryl hydrocarbon receptor antagonists
Abstract
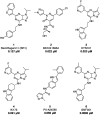
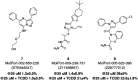
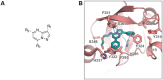
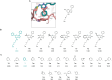
The aryl hydrocarbon receptor (AHR) is a versatile ligand-dependent transcription factor involved in diverse biological processes, from metabolic adaptations to immune system regulation. Recognising its pivotal role in cancer immunology, AHR has become a promising target for cancer therapy. Here we report the discovery and structure-activity relationship studies of novel AHR antagonists. The potential AHR antagonists were identified via homology model-based high-throughput virtual screening and were experimentally verified in a luciferase reporter gene assay. The identified pyrazolo[1,5-a]pyrimidine-based AHR antagonist 7 (IC50 = 650 nM) was systematically optimised to elucidate structure-activity relationships and reach low nanomolar AHR antagonistic potency (7a, IC50 = 31 nM). Overall, the findings presented here provide new starting points for AHR antagonist development and offer insightful information on AHR antagonist structure-activity relationships.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
LinkOut - more resources
Full Text Sources

