Escherichia coli in urban marine sediments: interpreting virulence, biofilm formation, halotolerance, and antibiotic resistance to infer contamination or naturalization
- PMID: 39246828
- PMCID: PMC11378635
- DOI: 10.1093/femsmc/xtae024
Escherichia coli in urban marine sediments: interpreting virulence, biofilm formation, halotolerance, and antibiotic resistance to infer contamination or naturalization
Abstract
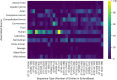
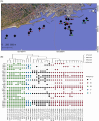
Marine sediments have been suggested as a reservoir for pathogenic bacteria, including Escherichia coli. The origins, and properties promoting survival of E. coli in marine sediments (including osmotolerance, biofilm formation capacity, and antibiotic resistance), have not been well-characterized. Phenotypes and genotypes of 37 E. coli isolates from coastal marine sediments were characterized. The isolates were diverse: 30 sequence types were identified that have been previously documented in humans, livestock, and other animals. Virulence genes were found in all isolates, with more virulence genes found in isolates sampled from sediment closer to the effluent discharge point of a wastewater treatment plant. Antibiotic resistance was demonstrated phenotypically for one isolate, which also carried tetracycline resistance genes on a plasmid. Biofilm formation capacity varied for the different isolates, with most biofilm formed by phylogroup B1 isolates. All isolates were halotolerant, growing at 3.5% NaCl. This suggests that the properties of some isolates may facilitate survival in marine environments and can explain in part how marine sediments can be a reservoir for pathogenic E. coli. As disturbance of sediment could resuspend bacteria, this should be considered as a potential contributor to compromised bathing water quality at nearby beaches.
Keywords: antibiotic resistance; bathing water; biofim; environmental Escherichia coli; halotolerance; whole genome sequencing.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
