Cross comparison of alternative diagnostic protocols including substitution to the clinical sample, RNA extraction method and nucleic acid amplification technology for COVID-19 diagnosis
- PMID: 39247206
- PMCID: PMC11377848
- DOI: 10.3389/fmolb.2024.1445142
Cross comparison of alternative diagnostic protocols including substitution to the clinical sample, RNA extraction method and nucleic acid amplification technology for COVID-19 diagnosis
Abstract
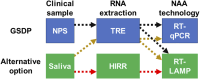
Background: the gold-standard diagnostic protocol (GSDP) for COVID-19 consists of a nasopharyngeal swab (NPS) sample processed through traditional RNA extraction (TRE) and amplified with retrotranscription quantitative polymerase chain reaction (RT-qPCR). Multiple alternatives were developed to decrease time/cost of GSDP, including alternative clinical samples, RNA extraction methods and nucleic acid amplification. Thus, we carried out a cross comparison of various alternatives methods against GSDP and each other.
Methods: we tested alternative diagnostic methods using saliva, heat-induced RNA release (HIRR) and a colorimetric retrotranscription loop-mediated isothermal amplification (RT-LAMP) as substitutions to the GSDP.
Results: RT-LAMP using NPS processed by TRE showed high sensitivity (96%) and specificity (97%), closely matching GSDP. When saliva was processed by TRE and amplified with both RT-LAMP and RT-qPCR, RT-LAMP yielded high diagnostic parameters (88%-96% sensitivity and 95%-100% specificity) compared to RT-qPCR. Nonetheless, when saliva processed by TRE and detected by RT-LAMP was compared against the GSDP, the resulting diagnostic values for sensitivity (78%) and specificity (87%) were somewhat high but still short of those of the GSDP. Finally, saliva processed with HIRR and detected via RT-LAMP was the simplest and fastest method, but its sensitivity against GSDP was too low (56%) for any clinical application. Also, in this last method, the acidity of a large percentage of saliva samples (9%-22%) affected the pH-sensitive colorimetric indicator used in the test, requiring the exclusion of these acidic samples or an extra step for pH correction.
Discussion: our comparison shows that RT-LAMP technology has diagnostic performance on par with RT-qPCR; likewise, saliva offers the same diagnostic functionality as NPS when subjected to a TRE method. Nonetheless, use of direct saliva after a HIRR and detected with RT-LAMP does not produce an acceptable diagnostic performance.
Keywords: COVID-19; RT-LAMP; SARS-CoV-2; alternative protocol; saliva sample.
Copyright © 2024 Segura-Ulate, Apú, Cortés, Querol-Audi, Zaldívar, Ortega, Flores-Mora, Gatica-Arias and Madrigal-Redondo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Baba M. M., Bitew M., Fokam J., Lelo E. A., Ahidjo A., Asmamaw K., et al. (2021). Diagnostic performance of a colorimetric RT -LAMP for the identification of SARS-CoV-2: a multicenter prospective clinical evaluation in sub-Saharan Africa. eClinicalMedicine 40, 101101. 10.1016/j.eclinm.2021.101101 - DOI - PMC - PubMed
-
- Babafemi E. O., Cherian B. P., Banting L., Mills G. A., Ngianga K. (2017). Effectiveness of real-time polymerase chain reaction assay for the detection of Mycobacterium tuberculosis in pathological samples: a systematic review and meta-analysis. Syst. Rev. 6 (1), 215. 10.1186/s13643-017-0608-2 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

