Vasculogenic mimicry-related gene prognostic index for predicting prognosis, immune microenvironment in clear cell renal cell carcinoma
- PMID: 39247316
- PMCID: PMC11380016
- DOI: 10.1016/j.heliyon.2024.e36235
Vasculogenic mimicry-related gene prognostic index for predicting prognosis, immune microenvironment in clear cell renal cell carcinoma
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) is a highly aggressive cancer associated with higher death rates. However, traditional anti-angiogenic therapies have limited effectiveness due to drug resistance. Vascular mimicry (VM) provides a different way for tumors to develop blood vessels without relying on endothelial cells or angiogenesis. However, the intricate mechanisms and interplay between it and the immune microenvironment in ccRCC remain unclear.
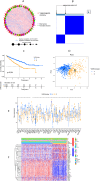
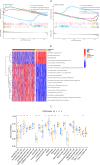
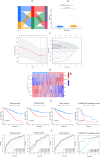
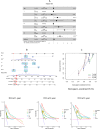
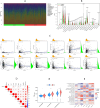
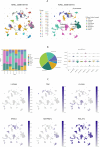
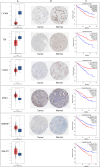
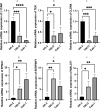
Methods: A PubMed and GeneCards literature review was conducted to identify VM-related genes (VMRGs). VMRGs expression profiles were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), developing a novel VM risk score model and nomogram for ccRCC. The EBI ArrayExpress database (the validation set) was obtained to validate the prognostic model. The relationship between VMRGs risk score clinical characteristics and immune infiltration was investigated. Finally, the expression of six model VMRGs was validated using single-cell analysis, GEPIA, Human Protein Atlas (HPA), and quantitative Real-time PCR (qRT-PCR).
Results: Cox regression analysis and nomogram identified L1CAM, TEK, CLDN4, EFNA1, SERPINF1, and MALAT1 as independent prognostic risk factors, which could be used to stratify the ccRCC population into two risk groups with distinct immune profiles and responsiveness to immunotherapy. The results of single-cell analysis, GEPIA, HPA, and qRT-PCR validated the model genes' expression.
Conclusions: Our novel findings constructed a convenient and reliable 6 gene signatures as potential immunologic and prognostic biomarkers of VM in ccRCC.
Keywords: Clear cell renal cell carcinoma; Immune infiltration; Nomogram; Tumor microenvironment; Vasculogenic mimicry.
© 2024 The Authors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

