Genome-Wide Mendelian Randomization Identifies Ferroptosis-Related Drug Targets for Alzheimer's Disease
- PMID: 39247875
- PMCID: PMC11380310
- DOI: 10.3233/ADR-240062
Genome-Wide Mendelian Randomization Identifies Ferroptosis-Related Drug Targets for Alzheimer's Disease
Abstract
Background: Alzheimer's disease (AD) currently lacks effective disease-modifying treatments. Recent research suggests that ferroptosis could be a potential therapeutic target. Mendelian randomization (MR) is a widely used method for identifying novel therapeutic targets.
Objective: Employ genetic information to evaluate the causal impact of ferroptosis-related genes on the risk of AD.
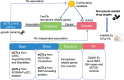
Methods: 564 ferroptosis-related genes were obtained from FerrDb. We derived genetic instrumental variables for these genes using four brain quantitative trait loci (QTL) and two blood QTL datasets. Summary-data-based Mendelian randomization (SMR) and two-sample MR methods were applied to estimate the causal effects of ferroptosis-related genes on AD. Using extern transcriptomic datasets and triple-transgenic mouse model of AD (3xTg-AD) to further validate the gene targets identified by the MR analysis.
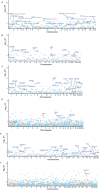
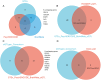
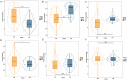
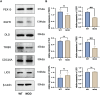
Results: We identified 17 potential AD risk gene targets from GTEx, 13 from PsychENCODE, and 22 from BrainMeta (SMR p < 0.05 and HEIDI test p > 0.05). Six overlapping ferroptosis-related genes associated with AD were identified, which could serve as potential therapeutic targets (PEX10, CDC25A, EGFR, DLD, LIG3, and TRIB3). Additionally, we further pinpointed risk genes or proteins at the blood tissue and pQTL levels. Notably, EGFR demonstrated significant dysregulation in the extern transcriptomic datasets and 3xTg-AD models.
Conclusions: This study provides genetic evidence supporting the potential therapeutic benefits of targeting the six druggable genes for AD treatment, especially for EGFR (validated by transcriptome and 3xTg-AD), which could be useful for prioritizing AD drug development in the field of ferroptosis.
Keywords: 3xTg-AD; Alzheimer’s disease; Mendelian randomization; ferroptosis.
© 2024 – The authors. Published by IOS Press.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures
Similar articles
-
Mendelian randomization analysis identifies druggable genes and drugs repurposing for chronic obstructive pulmonary disease.Front Cell Infect Microbiol. 2024 Apr 10;14:1386506. doi: 10.3389/fcimb.2024.1386506. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38660492 Free PMC article.
-
Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer's Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification.Curr Issues Mol Biol. 2025 Feb 11;47(2):118. doi: 10.3390/cimb47020118. Curr Issues Mol Biol. 2025. PMID: 39996839 Free PMC article.
-
Systematic druggable genome-wide Mendelian randomisation identifies therapeutic targets for Alzheimer's disease.J Neurol Neurosurg Psychiatry. 2023 Nov;94(11):954-961. doi: 10.1136/jnnp-2023-331142. Epub 2023 Jun 22. J Neurol Neurosurg Psychiatry. 2023. PMID: 37349091 Free PMC article.
-
Systematic druggable genome-wide Mendelian randomization identifies therapeutic targets for hyperemesis gravidarum.BMC Pregnancy Childbirth. 2024 Dec 23;24(1):848. doi: 10.1186/s12884-024-07077-8. BMC Pregnancy Childbirth. 2024. PMID: 39716115 Free PMC article.
-
The Potential of Ferroptosis-Targeting Therapies for Alzheimer's Disease: From Mechanism to Transcriptomic Analysis.Front Aging Neurosci. 2021 Dec 20;13:745046. doi: 10.3389/fnagi.2021.745046. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34987375 Free PMC article. Review.
Cited by
-
The role of ferroptosis in Alzheimer's disease: Mechanisms and therapeutic potential (Review).Mol Med Rep. 2025 Jul;32(1):192. doi: 10.3892/mmr.2025.13557. Epub 2025 May 9. Mol Med Rep. 2025. PMID: 40341407 Free PMC article. Review.
-
Exploration of the multiomics-based mechanisms of Gancao Nourishing-Yin decoction in regulating mitochondrial metabolic genes CYB5R3 and PICK1 to influence glioma progression.Discov Oncol. 2025 Apr 8;16(1):487. doi: 10.1007/s12672-025-02168-0. Discov Oncol. 2025. PMID: 40198489 Free PMC article.
References
-
- Jakaria M, Belaidi AA, Bush AI, et al.. Ferroptosis as a mechanism of neurodegeneration in Alzheimer’s disease. J Neurochem 2021; 159: 804–825. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

