The gap-free genome of Forsythia suspensa illuminates the intricate landscape of centromeres
- PMID: 39247880
- PMCID: PMC11374533
- DOI: 10.1093/hr/uhae185
The gap-free genome of Forsythia suspensa illuminates the intricate landscape of centromeres
Abstract
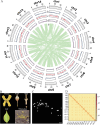
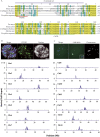
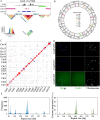
Forsythia suspensa, commonly known as weeping forsythia, holds significance in traditional medicine and horticulture. Despite its ecological and cultural importance, the existing reference genome presents challenges with duplications and gaps, hindering in-depth genomic analyses. Here, we present a Telomere-to-Telomere (T2T) assembly of the F. suspensa genome, integrating Oxford Nanopore Technologies (ONT) ultra-long, Hi-C datasets, and high-fidelity (HiFi) sequencing data. The T2T reference genome (Fsus-CHAU) consists of 14 chromosomes, totaling 688.79 Mb, and encompasses 33 932 predicted protein-coding genes. Additionally, we characterize functional centromeres in the F. suspensa genome by developing a specific CENH3 antibody. We demonstrate that centromeric regions in F. suspensa exhibit a diverse array of satellites, showcasing distinctive types with unconventional lengths across various chromosomes. This discovery offers implications for the adaptability of CENH3 and the potential influence on centromere dynamics. Furthermore, after assessing the insertion time of full-length LTRs within centromeric regions, we found that they are older compared to those across the entire genome, contrasting with observations in other species where centromeric retrotransposons are typically young. We hypothesize that asexual reproduction may impact retrotransposon dynamics, influencing centromere evolution. In conclusion, our T2T assembly of the F. suspensa genome, accompanied by detailed genomic annotations and centromere analysis, significantly enhances F. suspensa potential as a subject of study in fields ranging from ecology and horticulture to traditional medicine.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
No conflict of interest was declared.
Figures


References
-
- Liu Q, Chen X, Guo L. et al. Comparison of the growth adaptation of six Forthysia species in the quaternary red soil of South China. Res Soil Water Conserv. 2020;27:357–69
-
- Li Y, Wang F, Pei N. et al. The updated weeping forsythia genome reveals the genomic basis for the evolution and the forsythin and forsythoside a biosynthesis. Hortic Plant J. 2023;9:1149–61
-
- Liu C, Huang Y, Guo X. et al. Young retrotransposons and non-B DNA structures promote the establishment of dominant rye centromere in the 1RS.1BL fused centromere. New Phytol. 2024;241:607–22 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

