SlTrxh functions downstream of SlMYB86 and positively regulates nitrate stress tolerance via S-nitrosation in tomato seedling
- PMID: 39247888
- PMCID: PMC11374535
- DOI: 10.1093/hr/uhae184
SlTrxh functions downstream of SlMYB86 and positively regulates nitrate stress tolerance via S-nitrosation in tomato seedling
Abstract
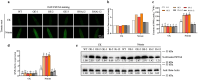
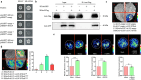
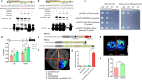
Nitric oxide (NO) is a redox-dependent signaling molecule that plays a crucial role in regulating a wide range of biological processes in plants. It functions by post-translationally modifying proteins, primarily through S-nitrosation. Thioredoxin (Trx), a small and ubiquitous protein with multifunctional properties, plays a pivotal role in the antioxidant defense system. However, the regulatory mechanism governing the response of tomato Trxh (SlTrxh) to excessive nitrate stress remains unknown. In this study, overexpression or silencing of SlTrxh in tomato led to increased or decreased nitrate stress tolerance, respectively. The overexpression of SlTrxh resulted in a reduction in levels of reactive oxygen species (ROS) and an increase in S-nitrosothiol (SNO) contents; conversely, silencing SlTrxh exhibited the opposite trend. The level of S-nitrosated SlTrxh was increased and decreased in SlTrxh overexpression and RNAi plants after nitrate treatment, respectively. SlTrxh was found to be susceptible to S-nitrosation both in vivo and in vitro, with Cysteine 54 potentially being the key site for S-nitrosation. Protein interaction assays revealed that SlTrxh physically interacts with SlGrx9, and this interaction is strengthened by S-nitrosation. Moreover, a combination of yeast one-hybrid (Y1H), electrophoretic mobility shift assay (EMSA), chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR), and transient expression assays confirmed the direct binding of SlMYB86 to the SlTrxh promoter, thereby enhancing its expression. SlMYB86 is located in the nucleus and SlMYB86 overexpressed and knockout tomato lines showed enhanced and decreased nitrate stress tolerance, respectively. Our findings indicate that SlTrxh functions downstream of SlMYB86 and highlight the potential significance of S-nitrosation of SlTrxh in modulating its function under nitrate stress.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
We declare the absence of any conflicts of interest.
Figures





References
-
- Tilman D, Cassman KG, Matson PA. et al. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–7 - PubMed
-
- Ju XT, Kou CL, Christie P. et al. Changes in the soil environment from excessive application of fertilizers and manures to two contrasting intensive cropping systems on the North China plain. Environ Pollut. 2007;145:497–506 - PubMed
-
- Yu L, Ma S, Zhang X. et al. Ancient rapid functional differentiation and fixation of the duplicated members in rice Dof genes after whole genome duplication. Plant J: Cell Mol Biol. 2021;108:1365–81 - PubMed
-
- Qin SQ, Quan Z, Ma J. et al. Regulating nitrate excess in lettuce-planted greenhouse soil with available carbon addition through irrigation. Environ Sci Pollut R. 2019;26:19241–9 - PubMed
-
- Xu HN, He XZ, Wang K. et al. Identification of early nitrate stress response genes in spinach roots by suppression subtractive hybridization. Plant Mol Biol Report. 2012;30:633–42
LinkOut - more resources
Full Text Sources

