DFF-ChIP: a method to detect and quantify complex interactions between RNA polymerase II, transcription factors, and chromatin
- PMID: 39248105
- PMCID: PMC11472042
- DOI: 10.1093/nar/gkae760
DFF-ChIP: a method to detect and quantify complex interactions between RNA polymerase II, transcription factors, and chromatin
Abstract
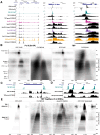
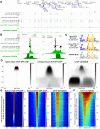
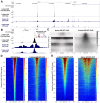
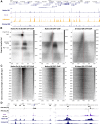
Recently, we introduced a chromatin immunoprecipitation (ChIP) technique utilizing the human DNA Fragmentation Factor (DFF) to digest the DNA prior to immunoprecipitation (DFF-ChIP) that provides the precise location of transcription complexes and their interactions with neighboring nucleosomes. Here we expand the technique to new targets and provide useful information concerning purification of DFF, digestion conditions, and the impact of crosslinking. DFF-ChIP analysis was performed individually for subunits of Mediator, DSIF, and NELF that that do not interact with DNA directly, but rather interact with RNA polymerase II (Pol II). We found that Mediator was associated almost exclusively with preinitiation complexes (PICs). DSIF and NELF were associated with engaged Pol II and, in addition, potential intermediates between PICs and early initiation complexes. DFF-ChIP was then used to analyze the occupancy of a tight binding transcription factor, CTCF, and a much weaker binding factor, glucocorticoid receptor (GR), with and without crosslinking. These results were compared to those from standard ChIP-Seq that employs sonication and to CUT&RUN which utilizes MNase to fragment the genomic DNA. Our findings indicate that DFF-ChIP reveals details of occupancy that are not available using other methods including information revealing pertinent protein:protein interactions.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

