Making target sites in large structured RNAs accessible to RNA-cleaving DNAzymes through hybridization with synthetic DNA oligonucleotides
- PMID: 39248110
- PMCID: PMC11472044
- DOI: 10.1093/nar/gkae778
Making target sites in large structured RNAs accessible to RNA-cleaving DNAzymes through hybridization with synthetic DNA oligonucleotides
Abstract
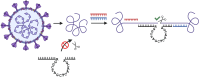
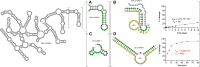
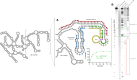
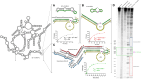
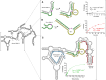
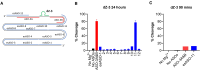
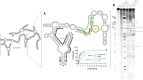
The 10-23 DNAzyme is one of the most active DNA-based enzymes, and in theory, can be designed to target any purine-pyrimidine junction within an RNA sequence for cleavage. However, purine-pyrimidine junctions within a large, structured RNA (lsRNA) molecule of biological origin are not always accessible to 10-23, negating its general utility as an RNA-cutting molecular scissor. Herein, we report a generalizable strategy that allows 10-23 to access any purine-pyrimidine junction within an lsRNA. Using three large SARS-CoV-2 mRNA sequences of 566, 584 and 831 nucleotides in length as model systems, we show that the use of antisense DNA oligonucleotides (ASOs) that target the upstream and downstream regions flanking the cleavage site can restore the activity (kobs) of previously poorly active 10-23 DNAzyme systems by up to 2000-fold. We corroborated these findings mechanistically using in-line probing to demonstrate that ASOs reduced 10-23 DNAzyme target site structure within the lsRNA substrates. This approach represents a simple, efficient, cost-effective, and generalizable way to improve the accessibility of 10-23 to a chosen target site within an lsRNA molecule, especially where direct access to the genomic RNA target is necessary.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Yan J., Ran M., Shen X., Zhang H.. Therapeutic DNAzymes: from structure design to clinical applications. Adv. Mater. 2023; 35:2300374. - PubMed
-
- Cozma I., McConnell E.M., Brennan J.D., Li Y.. DNAzymes as key components of biosensing systems for the detection of biological targets. Biosens. Bioelectron. 2021; 177:112972. - PubMed
-
- McConnell E.M., Cozma I., Morrison D., Li Y.. Biosensors made of synthetic functional nucleic acids toward better Human health. Anal. Chem. 2020; 92:327–344. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

