Heat shock transcription factor 1 facilitates liver cancer progression by driving super-enhancer-mediated transcription of MYCN
- PMID: 39248163
- PMCID: PMC11382014
- DOI: 10.1002/cam4.70157
Heat shock transcription factor 1 facilitates liver cancer progression by driving super-enhancer-mediated transcription of MYCN
Abstract
Background: Heat shock transcription factors (HSFs) play crucial roles in the development of malignancies. However, the specific roles of HSFs in hepatocellular carcinoma (HCC) have yet to be fully elucidated.
Aims: To explore the involvement of the HSF family, particularly HSF1, in the progression and prognosis of HCC.
Materials & methods: We conducted a thorough analysis of HSF expression and copy number variations across various cancer datasets. Specifically focusing on HSF1, we examined its expression levels and prognostic implications in HCC. In vitro and in vivo experiments were carried out to evaluate the impact of HSF1 on liver cancer cell proliferation. Additionally, we utilized CUT&Tag, H3K27 acetylation enrichment, and RNA sequencing (RNA-seq) to investigate the super-enhancer (SE) regulatory landscapes of HSF1 in liver cancer cell lines.
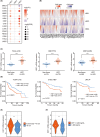
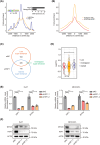
Results: HSF1 expression is elevated in HCC and is linked to poor prognosis in several datasets. HSF1 stimulates liver cancer cell proliferation both in vitro and in vivo, partly through modulation of H3K27ac levels, influencing enhancer distribution. Mechanistically, our findings demonstrate that HSF1 transcriptionally activates MYCN expression by binding to its promoter and SE elements, thereby promoting liver cancer cell proliferation. Moreover, increased MYCN expression was detected in HCC tumors and correlated with unfavorable patient outcomes.
Discussion: Our study sheds light on previously unexplored aspects of HSF1 biology, identifying it as a transcription factor capable of shaping the epigenetic landscape in the context of HCC. Given HSF1's potential as an epigenetic regulator, targeting the HSF1-MYCN axis could open up new therapeutic possibilities for HCC treatment.
Conclusion: The HSF1-MYCN axis constitutes a transcription-dependent regulatory mechanism that may function as both a prognostic indicator and a promising therapeutic target in liver cancer. Further exploration of this axis could yield valuable insights into novel treatment strategies for HCC.
Keywords: HSF1; MYCN; liver cancer; super‐enhancer; transcriptional regulation.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular Carcinoma. Nat Rev Dis Prim. 2021;7:6. - PubMed
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. - PubMed
-
- Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151‐172. - PubMed
-
- Wang Z, Wang Y, Gao P, Ding J. Immune checkpoint inhibitor resistance in hepatocellular carcinoma. Cancer Lett. 2023;555:216038. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

