Efflux pumps mediate changes to fundamental bacterial physiology via membrane potential
- PMID: 39248573
- PMCID: PMC11481890
- DOI: 10.1128/mbio.02370-24
Efflux pumps mediate changes to fundamental bacterial physiology via membrane potential
Abstract
Efflux pumps are well known to be an important mechanism for removing noxious substances such as antibiotics from bacteria. Given that many antibiotics function by accumulating inside bacteria, efflux pumps contribute to resistance. Efflux pump inactivation is a potential strategy to combat antimicrobial resistance, as bacteria would not be able to pump out antibiotics. We recently discovered that the impact of loss of efflux function is only apparent in actively growing cells. We demonstrated that the global transcriptome of Salmonella Typhimurium is drastically altered during slower growth leading to stationary-phase cells having a remodeled, less permeable envelope that prevents antibiotics entering the cell. Here, we investigated the effects of deleting the major efflux pump of Salmonella Typhimurium, AcrB, on global gene transcription across growth. We revealed that an acrB knockout entered stationary phase later than the wild-type strain SL1344 and displayed increased and prolonged expression of genes responsible for anaerobic energy metabolism. We devised a model linking efflux and membrane potential, whereby deactivation of AcrB prevents influx of protons across the inner membrane and thereby hyperpolarization. Knockout or deactivation of AcrB was demonstrated to increase membrane potential. We propose that the global transcription regulator ArcBA senses changes to the redox state of the quinol pool (linked to the membrane potential of the bacterium) and coordinates the shift from exponential to stationary phase via the key master regulators RpoS, Rsd, and Rmf. Inactivation of efflux pumps therefore influences the fundamental physiology of Salmonella, with likely impacts on multiple phenotypes.IMPORTANCEWe demonstrate for the first time that deactivation of efflux pumps brings about changes to gross bacterial physiology and metabolism. Rather than simply being a response to noxious substances, efflux pumps appear to play a key role in maintenance of membrane potential and thereby energy metabolism. This discovery suggests that efflux pump inhibition or inactivation might have unforeseen positive consequences on antibiotic activity. Given that stationary-phase bacteria are more resistant to antibiotic uptake, late entry into stationary phase would prolong antibiotic accumulation by bacteria. Furthermore, membrane hyperpolarization could result in increased generation of reactive species proposed to be important for the activity of some antibiotics. Finally, changes in gross physiology could also explain the decreased virulence of efflux mutants.
Keywords: AcrAB-TolC; ArcBA; RND efflux pump; energy metabolism; hyperpolarization; redox state.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
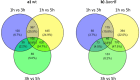




References
-
- Webber MA, Bailey AM, Blair JMA, Morgan E, Stevens MP, Hinton JCD, Ivens A, Wain J, Piddock LJV. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol 191:4276–4285. doi:10.1128/JB.00363-09 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- 108876/B/15/Z/Wellcome Trust (WT)
- MIBTP studentship/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/M02623X/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- Centre for Systems Modelling and Quantitative Biomedicine,Research Development Fund/University of Birmingham
LinkOut - more resources
Full Text Sources
Medical
