Rifampicin tolerance and growth fitness among isoniazid-resistant clinical Mycobacterium tuberculosis isolates from a longitudinal study
- PMID: 39250422
- PMCID: PMC11383526
- DOI: 10.7554/eLife.93243
Rifampicin tolerance and growth fitness among isoniazid-resistant clinical Mycobacterium tuberculosis isolates from a longitudinal study
Abstract
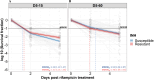
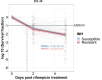
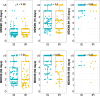
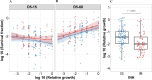
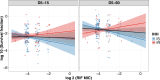
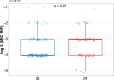
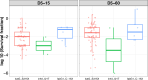
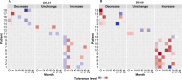
Antibiotic tolerance in Mycobacterium tuberculosis reduces bacterial killing, worsens treatment outcomes, and contributes to resistance. We studied rifampicin tolerance in isolates with or without isoniazid resistance (IR). Using a minimum duration of killing assay, we measured rifampicin survival in isoniazid-susceptible (IS, n=119) and resistant (IR, n=84) isolates, correlating tolerance with bacterial growth, rifampicin minimum inhibitory concentrations (MICs), and isoniazid-resistant mutations. Longitudinal IR isolates were analyzed for changes in rifampicin tolerance and genetic variant emergence. The median time for rifampicin to reduce the bacterial population by 90% (MDK90) increased from 1.23 days (IS) and 1.31 days (IR) to 2.55 days (IS) and 1.98 days (IR) over 15-60 days of incubation, indicating fast and slow-growing tolerant sub-populations. A 6 log10-fold survival fraction classified tolerance as low, medium, or high, showing that IR is linked to increased tolerance and faster growth (OR = 2.68 for low vs. medium, OR = 4.42 for low vs. high, p-trend = 0.0003). High tolerance in IR isolates was associated with rifampicin treatment in patients and genetic microvariants. These findings suggest that IR tuberculosis should be assessed for high rifampicin tolerance to optimize treatment and prevent the development of multi-drug-resistant tuberculosis.
Keywords: infectious disease; microbiology; mycobacterium; rifampicin; tolerance; tuberculosis.
© 2024, Vijay, Bao et al.
Conflict of interest statement
SV, NB, DV, LN, DT, NQ, LT, HN, VH, PT, DH, NL, MC, GT, BJ, NT No competing interests declared
Figures






Update of
-
Rifampicin tolerance and growth fitness among isoniazid-resistant clinical Mycobacterium tuberculosis isolates: an in-vitro longitudinal study.bioRxiv [Preprint]. 2024 May 4:2023.11.22.568240. doi: 10.1101/2023.11.22.568240. bioRxiv. 2024. Update in: Elife. 2024 Sep 09;13:RP93243. doi: 10.7554/eLife.93243. PMID: 38045287 Free PMC article. Updated. Preprint.
References
-
- Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TEA, Crook DW, Iqbal Z. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nature Communications. 2015;6:10063. doi: 10.1038/ncomms10063. - DOI - PMC - PubMed

