Engineered NLS-chimera downregulates expression of aggregation-prone endogenous FUS
- PMID: 39251571
- PMCID: PMC11384663
- DOI: 10.1038/s41467-024-52151-6
Engineered NLS-chimera downregulates expression of aggregation-prone endogenous FUS
Abstract
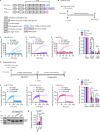
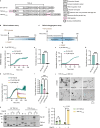
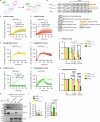
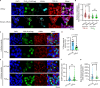
Importin β-superfamily nuclear import receptors (NIRs) mitigate mislocalization and aggregation of RNA-binding proteins (RBPs), like FUS and TDP-43, which are implicated in neurodegenerative diseases. NIRs potently disaggregate RBPs by recognizing their nuclear localization signal (NLS). However, disease-causing mutations in NLS compromise NIR binding and activity. Here, we define features that characterize the anti-aggregation activity of NIR and NLS. We find that high binding affinity between NIR and NLS, and optimal NLS location relative to the aggregating domain plays a role in determining NIR disaggregation activity. A designed FUS chimera (FUSIBB), carrying the importin β binding (IBB) domain, is solubilized by importin β in vitro, translocated to the nucleus in cultured cells, and downregulates the expression of endogenous FUS. In this study, we posit that guiding the mutual recognition of NLSs and NIRs will aid the development of therapeutics, illustrated by the highly soluble FUSIBB replacing the aggregation-prone endogenous FUS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R35GM138109/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- RF1 NS121143/NS/NINDS NIH HHS/United States
- R21 NS128396/NS/NINDS NIH HHS/United States
- T32 GM144302/GM/NIGMS NIH HHS/United States
- R01 NS121143/NS/NINDS NIH HHS/United States
- R35 GM140733/GM/NIGMS NIH HHS/United States
- R35GM140733/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- RF1NS121143/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R35 GM138109/GM/NIGMS NIH HHS/United States
- R21NS128396/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
LinkOut - more resources
Full Text Sources
Miscellaneous

