FoxO transcription factors actuate the formative pluripotency specific gene expression programme
- PMID: 39251582
- PMCID: PMC11384738
- DOI: 10.1038/s41467-024-51794-9
FoxO transcription factors actuate the formative pluripotency specific gene expression programme
Abstract
Naïve pluripotency is sustained by a self-reinforcing gene regulatory network (GRN) comprising core and naïve pluripotency-specific transcription factors (TFs). Upon exiting naïve pluripotency, embryonic stem cells (ESCs) transition through a formative post-implantation-like pluripotent state, where they acquire competence for lineage choice. However, the mechanisms underlying disengagement from the naïve GRN and initiation of the formative GRN are unclear. Here, we demonstrate that phosphorylated AKT acts as a gatekeeper that prevents nuclear localisation of FoxO TFs in naïve ESCs. PTEN-mediated reduction of AKT activity upon exit from naïve pluripotency allows nuclear entry of FoxO TFs, enforcing a cell fate transition by binding and activating formative pluripotency-specific enhancers. Indeed, FoxO TFs are necessary and sufficient for the activation of the formative pluripotency-specific GRN. Our work uncovers a pivotal role for FoxO TFs in establishing formative post-implantation pluripotency, a critical early embryonic cell fate transition.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

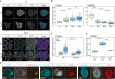




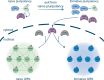
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- I 5958/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- DOC 72/FWF_/Austrian Science Fund FWF/Austria
- W 1261/Austrian Science Fund (Fonds zur Förderung der Wissenschaftlichen Forschung)
- VRG14-006/Vienna Science and Technology Fund (Wiener Wissenschafts-, Forschungs- und Technologiefonds)
- DOC/25622/Österreichischen Akademie der Wissenschaften (Austrian Academy of Sciences)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

