Long-term longitudinal analysis of 4,187 participants reveals insights into determinants of clonal hematopoiesis
- PMID: 39251642
- PMCID: PMC11385577
- DOI: 10.1038/s41467-024-52302-9
Long-term longitudinal analysis of 4,187 participants reveals insights into determinants of clonal hematopoiesis
Abstract
Clonal hematopoiesis of indeterminate potential (CHIP) is linked to diverse aging-related diseases, including hematologic malignancy and atherosclerotic cardiovascular disease (ASCVD). While CHIP is common among older adults, the underlying factors driving its development are largely unknown. To address this, we performed whole-exome sequencing on 8,374 blood DNA samples collected from 4,187 Atherosclerosis Risk in Communities Study (ARIC) participants over a median follow-up of 21 years. During this period, 735 participants developed incident CHIP. Splicing factor genes (SF3B1, SRSF2, U2AF1, and ZRSR2) and TET2 CHIP grow significantly faster than DNMT3A non-R882 clones. We find that age at baseline and sex significantly influence the incidence of CHIP, while ASCVD and other traditional ASCVD risk factors do not exhibit such associations. Additionally, baseline synonymous passenger mutations are strongly associated with CHIP status and are predictive of new CHIP clone acquisition and clonal growth over extended follow-up, providing valuable insights into clonal dynamics of aging hematopoietic stem and progenitor cells. This study also reveals associations between germline genetic variants and incident CHIP. Our comprehensive longitudinal assessment yields insights into cell-intrinsic and -extrinsic factors contributing to the development and progression of CHIP clones in older adults.
© 2024. The Author(s).
Conflict of interest statement
M.C.H. reports research grants from Genentech, advisory board service for Miga Health, and consulting fees from Comanche Biopharma, all unrelated to the present work. P.L. is an unpaid consultant to/or involved in clinical trials for Amgen, Baim Institute, Beren Therapeutics, Esperion Therapeutics, Genentech, Kancera, Kowa Pharmaceuticals, Novo Nordisk, Novartis, Sanofi-Regeneron. P.L. is a member of the scientific advisory board for Amgen, Caristo Diagnostics, CSL Behring, DalCor Pharmaceuticals, Dewpoint Therapeutics, Eulicid Bioimaging, Kancera, Kowa Pharmaceuticals, Olatec Therapeutics, MedImmune, Novartis, PlaqueTec, Polygon Therapeutics, TenSixteen Bio, Soley Therapeutics, and XBiotech, Inc. P.L.’s laboratory has received research funding in the last 2 years from Novartis, Novo Nordisk, and Genentech. P.L. is on the Board of Directors of XBiotech, Inc. P.L. has a financial interest in Xbiotech, a company developing therapeutic human antibodies, in TenSixteen Bio, a company targeting somatic mosaicism and clonal hematopoiesis of indeterminate potential (CHIP) to discover and develop novel therapeutics to treat age-related diseases, and in Soley Therapeutics, a biotechnology company that is combining artificial intelligence with molecular and cellular response detection for discovering and developing new drugs, currently focusing on cancer therapeutics. P.L.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Mass General Brigham in accordance with their conflict-of-interest policies. B.L.E. has received research funding from Novartis and Calico. He has received consulting fees from Abbvie. He is a member of the scientific advisory board and shareholder for Neomorph Inc., TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics, all distinct from the present work. P.N., A.G.B., and B.L.E. are scientific co-founders of TenSixteen Bio. P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, Astra Zeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Esperion Therapeutics, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, Novartis, TenSixteen Bio, and Tourmaline Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. C.M.B. reports grant/research support from Abbott Diagnostic, Akcea, Amgen, Arrowhead, Esperion, Ionis, Merck, New Amsterdam, Novartis, Novo Nordisk, Regeneron, Roche Diagnostic, NIH, AHA, ADA, consultation fees from Abbott Diagnostics, Alnylam Pharmaceuticals, Althera, Amarin, Amgen, Arrowhead, Astra Zeneca, Denka Seiken, Esperion, Genentech, Gilead, Illumina, Ionis, Matinas BioPharma Inc, Merck, New Amsterdam, Novartis, Novo Nordisk, Pfizer, Regeneron, Roche Diagnostic, TenSixteen Bio. The remaining authors declare no competing interests.
Figures
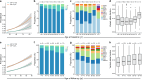

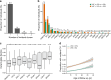



Update of
-
Long-term longitudinal analysis of 4,187 participants reveals new insights into determinants of incident clonal hematopoiesis.medRxiv [Preprint]. 2023 Sep 7:2023.09.05.23295093. doi: 10.1101/2023.09.05.23295093. medRxiv. 2023. Update in: Nat Commun. 2024 Sep 9;15(1):7858. doi: 10.1038/s41467-024-52302-9. PMID: 37732181 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- DP5 OD029586/OD/NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- R01 HL082945/HL/NHLBI NIH HHS/United States
- R01HL148050/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- T32 HL139425/HL/NHLBI NIH HHS/United States
- TNE-18CVD04/Fondation Leducq
- R01 HL087641/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- K08 HL166687/HL/NHLBI NIH HHS/United States
- R01 HL163099/HL/NHLBI NIH HHS/United States
- P50 CA206963/CA/NCI NIH HHS/United States
- R01 HL134892/HL/NHLBI NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 HL151627/HL/NHLBI NIH HHS/United States
- R01 HL166538/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- P01 CA108631/CA/NCI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- R01 AG063839/AG/NIA NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- R01 HL157073/HL/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- R01 HL148050/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

