Development of a prognostic model for NSCLC based on differential genes in tumour stem cells
- PMID: 39251710
- PMCID: PMC11383933
- DOI: 10.1038/s41598-024-71317-2
Development of a prognostic model for NSCLC based on differential genes in tumour stem cells
Abstract
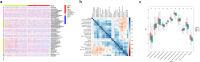
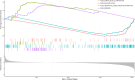
Non-small cell lung cancer (NSCLC) constitutes a significant portion of lung cancers and cytotoxic drugs (e.g. cisplatin) are currently the first-line treatment. However, NSCLC has developed resistance to this drug, which limits the therapeutic effect and thus affects prognosis. NSCLC sc-RNA-seq data were downloaded from the GEO database and Ku Leuven Laboratory for Functional Epigenetics, and bulk RNA-seq data were obtained from the TCGA database. The "Seurat" package was employed for scRNA-seq data processing, and the uniform manifold approximation and projection (UMAP) were applied for downscaling and cluster identification. Use the FindAllMarkers function to find differential genes (DEGs) for tumor stem cells. Then, we performed univariate regression analyses on the DEGs to identify potential prognostic genes. We created a machine learning framework based on potential prognostic genes, which combines 10 machine learning methods and their 101 combinations to get the optimal prognostic risk model. The model was evaluated in the training set and validation set. A nomogram was developed to provide physicians with a quantitative tool for prognosis prediction. Finally, we evaluated the expression and functionality of SLC2A1. We discovered 22 cell clusters containing 218379 cells by examining single-cell RNA sequencing datasets (GSE148071, KU_lom, GSE131907, GSE136246, GSE127465). Tumour cells were isolated for subpopulation analysis and 162 differential genes from SOX2_cancer were obtained. After univariate Cox analysis, we found 23 genes with prognostic potential prognostic value and utilized them to develop 101‑combination machine learning computational framework. We eventually picked the best performing 'StepCox[both] + RSF', which includes 8 genes. The model has a relatively high prediction accuracy in both TCGA and GEO datasets. In in vitro investigations, targeted suppression of the SLC2A1 gene resulted in significant reductions in proliferation, invasion and migration in A549 cells. In addition, a significant reduction in cisplatin resistance was seen in A549/DDP cells. The outcomes demonstrated the precision and credibility of the prognostic model for NSCLC, highlighting its potential significance in the treatment and prognosis of individuals affected by this disease. SLC2A1 may become a promising prognostic marker and a potential therapeutic target, offering valuable insights to inform clinical treatment decisions.
Keywords: Machine learning; NSCLC; Prognosis; Single cell.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






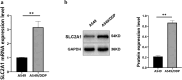


Similar articles
-
Comprehensive analysis of scRNA-Seq and bulk RNA-Seq reveals dynamic changes in the tumor immune microenvironment of bladder cancer and establishes a prognostic model.J Transl Med. 2023 Mar 27;21(1):223. doi: 10.1186/s12967-023-04056-z. J Transl Med. 2023. PMID: 36973787 Free PMC article.
-
Identifying lncRNAs and mRNAs related to survival of NSCLC based on bioinformatic analysis and machine learning.Aging (Albany NY). 2024 May 1;16(9):7799-7817. doi: 10.18632/aging.205783. Epub 2024 May 1. Aging (Albany NY). 2024. PMID: 38696317 Free PMC article.
-
Machine learning models reveal ARHGAP11A's impact on lymph node metastasis and stemness in NSCLC.Biofactors. 2025 Jan-Feb;51(1):e2141. doi: 10.1002/biof.2141. Epub 2024 Oct 31. Biofactors. 2025. PMID: 39482272
-
Construction of a novel radioresistance-related signature for prediction of prognosis, immune microenvironment and anti-tumour drug sensitivity in non-small cell lung cancer.Ann Med. 2025 Dec;57(1):2447930. doi: 10.1080/07853890.2024.2447930. Epub 2025 Jan 10. Ann Med. 2025. PMID: 39797413 Free PMC article.
-
Decoding temporal heterogeneity in NSCLC through machine learning and prognostic model construction.World J Surg Oncol. 2024 Jun 13;22(1):156. doi: 10.1186/s12957-024-03435-0. World J Surg Oncol. 2024. PMID: 38872167 Free PMC article.
Cited by
-
Machine learning-based model identifies a novel cuproptosis-related mitochondrial gene signature with a key role in the prognosis and treatment of lung adenocarcinoma.Oncol Lett. 2025 Aug 21;30(5):494. doi: 10.3892/ol.2025.15240. eCollection 2025 Nov. Oncol Lett. 2025. PMID: 40904599 Free PMC article.
-
C/EBPβ Regulates HIF-1α-Driven Invasion of Non-Small-Cell Lung Cancer Cells.Biomolecules. 2024 Dec 30;15(1):36. doi: 10.3390/biom15010036. Biomolecules. 2024. PMID: 39858431 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

