ALDH1A3 is the switch that determines the balance of ALDH+ and CD24-CD44+ cancer stem cells, EMT-MET, and glucose metabolism in breast cancer
- PMID: 39251846
- PMCID: PMC11493680
- DOI: 10.1038/s41388-024-03156-4
ALDH1A3 is the switch that determines the balance of ALDH+ and CD24-CD44+ cancer stem cells, EMT-MET, and glucose metabolism in breast cancer
Abstract
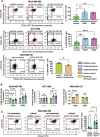
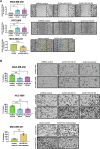
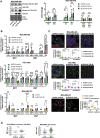
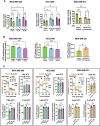
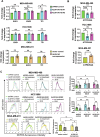
Plasticity is an inherent feature of cancer stem cells (CSCs) and regulates the balance of key processes required at different stages of breast cancer progression, including epithelial-to-mesenchymal transition (EMT) versus mesenchymal-to-epithelial transition (MET), and glycolysis versus oxidative phosphorylation. Understanding the key factors that regulate the switch between these processes could lead to novel therapeutic strategies that limit tumor progression. We found that aldehyde dehydrogenase 1A3 (ALDH1A3) regulates these cancer-promoting processes and the abundance of the two distinct breast CSC populations defined by high ALDH activity and CD24-CD44+ cell surface expression. While ALDH1A3 increases ALDH+ breast cancer cells, it inversely suppresses the CD24-CD44+ population by retinoic acid signaling-mediated gene expression changes. This switch in CSC populations induced by ALDH1A3 was paired with decreased migration but increased invasion and an intermediate EMT phenotype. We also demonstrate that ALDH1A3 increases oxidative phosphorylation and decreases glycolysis and reactive oxygen species (ROS). The effects of ALDH1A3 reduction were countered with the glycolysis inhibitor 2-deoxy-D-glucose (2DG). In cell culture and tumor xenograft models, 2DG suppresses the increase in the CD24-CD44+ population and ROS induced by ALDH1A3 knockdown. Combined inhibition of ALDH1A3 and glycolysis best reduces breast tumor growth and tumor-initiating cells, suggesting that the combination of targeting ALDH1A3 and glycolysis has therapeutic potential for limiting CSCs and tumor progression. Together, these findings identify ALDH1A3 as a key regulator of processes required for breast cancer progression and depletion of ALDH1A3 makes breast cancer cells more susceptible to glycolysis inhibition.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Frank NY, Frank MH. Cancer stem cells. Access Sci. 2024. 10.1036/1097-8542.105740.
-
- Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012;22:187–93. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

