Genome-centric metagenomes unveiling the hidden resistome in an anchialine cave
- PMID: 39252078
- PMCID: PMC11386340
- DOI: 10.1186/s40793-024-00612-2
Genome-centric metagenomes unveiling the hidden resistome in an anchialine cave
Abstract
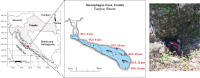
Background: Antibiotic resistance is a critical global concern, posing significant challenges to human health and medical treatments. Studying antibiotic resistance genes (ARGs) is essential not only in clinical settings but also in diverse environmental contexts. However, ARGs in unique environments such as anchialine caves, which connect both fresh and marine water, remain largely unexplored despite their intriguing ecological characteristics.
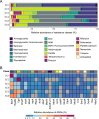
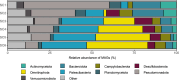
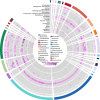
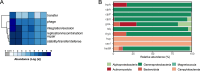
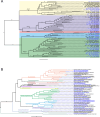
Results: We present the first study that comprehensively explores the occurrence and distribution of ARGs and mobile genetic elements (MGEs) within an anchialine cave. Utilizing metagenomic sequencing we uncovered a wide array of ARGs with the bacitracin resistance gene, bacA and multidrug resistance genes, being the most dominant. The cave's microbial community and associated resistome were significantly influenced by the salinity gradient. The discovery of novel β-lactamase variants revealed the cave's potential as a reservoir for previously undetected resistance genes. ARGs in the cave demonstrated horizontal transfer potential via plasmids, unveiling ecological implications.
Conclusions: These findings highlight the need for further exploration of the resistome in unique environments like anchialine caves. The interconnected dynamics of ARGs and MGEs within anchialine caves offer valuable insights into potential reservoirs and mechanisms of antibiotic resistance in natural ecosystems. This study not only advances our fundamental understanding but also highlights the need for a comprehensive approach to address antibiotic resistance in diverse ecological settings.
Keywords: Anchialine cave; Antibiotic resistance gene; Metagenomics; Mobile genetic element; Resistome; Subterranean estuary.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Metagenomic Insights into the Diverse Antibiotic Resistome of Non-Migratory Corvidae Species on the Qinghai-Tibetan Plateau.Vet Sci. 2025 Mar 23;12(4):297. doi: 10.3390/vetsci12040297. Vet Sci. 2025. PMID: 40284799 Free PMC article.
-
Antibiotic resistome alteration along a full-scale drinking water supply system deciphered by metagenome assembly: Regulated by seasonality, mobile gene elements and antibiotic resistant gene hosts.Sci Total Environ. 2023 Mar 1;862:160887. doi: 10.1016/j.scitotenv.2022.160887. Epub 2022 Dec 12. Sci Total Environ. 2023. PMID: 36521611
-
Multiplexed Target Enrichment Enables Efficient and In-Depth Analysis of Antimicrobial Resistome in Metagenomes.Microbiol Spectr. 2022 Dec 21;10(6):e0229722. doi: 10.1128/spectrum.02297-22. Epub 2022 Oct 26. Microbiol Spectr. 2022. PMID: 36287061 Free PMC article.
-
Profiles of Microbial Community and Antibiotic Resistome in Wild Tick Species.mSystems. 2022 Aug 30;7(4):e0003722. doi: 10.1128/msystems.00037-22. Epub 2022 Aug 1. mSystems. 2022. PMID: 35913190 Free PMC article. Review.
-
The challenges of defining the human nasopharyngeal resistome.Trends Microbiol. 2023 Aug;31(8):816-831. doi: 10.1016/j.tim.2023.02.008. Epub 2023 Mar 24. Trends Microbiol. 2023. PMID: 36967247 Review.
References
-
- World Health Organization. Global antimicrobial resistance and use surveillance system (GLASS) Report 2022. Braz Dent J. 2022.
Grants and funding
- KK.01.2.1.02.0335/European Union through the European Regional Development Fund
- KK.01.1.1.01.0003/European Union through the European Regional Development Fund
- KK.01.2.1.02.0335/European Union through the European Regional Development Fund
- CN_00000033/European Union - NextGenerationEU
- CN_00000033/European Union - NextGenerationEU
LinkOut - more resources
Full Text Sources
