This is a preprint.
Biological Insights from Schizophrenia-associated Loci in Ancestral Populations
- PMID: 39252912
- PMCID: PMC11383513
- DOI: 10.1101/2024.08.27.24312631
Biological Insights from Schizophrenia-associated Loci in Ancestral Populations
Abstract
Large-scale genome-wide association studies of schizophrenia have uncovered hundreds of associated loci but with extremely limited representation of African diaspora populations. We surveyed electronic health records of 200,000 individuals of African ancestry in the Million Veteran and All of Us Research Programs, and, coupled with genotype-level data from four case-control studies, realized a combined sample size of 13,012 affected and 54,266 unaffected persons. Three genome-wide significant signals - near PLXNA4, PMAIP1, and TRPA1 - are the first to be independently identified in populations of predominantly African ancestry. Joint analyses of African, European, and East Asian ancestries across 86,981 cases and 303,771 controls, yielded 376 distinct autosomal loci, which were refined to 708 putatively causal variants via multi-ancestry fine-mapping. Utilizing single-cell functional genomic data from human brain tissue and two complementary approaches, transcriptome-wide association studies and enhancer-promoter contact mapping, we identified a consensus set of 94 genes across ancestries and pinpointed the specific cell types in which they act. We identified reproducible associations of schizophrenia polygenic risk scores with schizophrenia diagnoses and a range of other mental and physical health problems. Our study addresses a longstanding gap in the generalizability of research findings for schizophrenia across ancestral populations, underlining shared biological underpinnings of schizophrenia across global populations in the presence of broadly divergent risk allele frequencies.
Figures

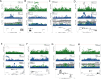








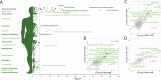
References
-
- Nagendra A. et al. Neighborhood socioeconomic status and racial disparities in schizophrenia: An exploration of domains of functioning. Schizophr. Res. 224, 95–101 (2020). - PubMed
-
- Anglin D. M. Racism and Social Determinants of Psychosis. Annu. Rev. Clin. Psychol. 19, 277–302 (2023). - PubMed
-
- Ikeda M. et al. Genome-wide association study of schizophrenia in a Japanese population. Biol. Psychiatry 69, 472–478 (2011). - PubMed
Publication types
Grants and funding
- R01 MH061675/MH/NIMH NIH HHS/United States
- U01 MH116442/MH/NIMH NIH HHS/United States
- R01 MH104964/MH/NIMH NIH HHS/United States
- U01 MH046276/MH/NIMH NIH HHS/United States
- R01 MH086135/MH/NIMH NIH HHS/United States
- I01 BX004189/BX/BLRD VA/United States
- R01 MH067257/MH/NIMH NIH HHS/United States
- U01 MH079470/MH/NIMH NIH HHS/United States
- R01 MH085548/MH/NIMH NIH HHS/United States
- R01 MH059586/MH/NIMH NIH HHS/United States
- U01 MH046289/MH/NIMH NIH HHS/United States
- R01 MH065588/MH/NIMH NIH HHS/United States
- K08 MH122911/MH/NIMH NIH HHS/United States
- R01 MH065571/MH/NIMH NIH HHS/United States
- U01 MH079469/MH/NIMH NIH HHS/United States
- R01 MH060870/MH/NIMH NIH HHS/United States
- R01 MH081800/MH/NIMH NIH HHS/United States
- R01 MH065558/MH/NIMH NIH HHS/United States
- R01 MH059571/MH/NIMH NIH HHS/United States
- R01 MH094320/MH/NIMH NIH HHS/United States
- R01 MH059565/MH/NIMH NIH HHS/United States
- R01 MH065554/MH/NIMH NIH HHS/United States
- R01 MH065578/MH/NIMH NIH HHS/United States
- R01 AG067025/AG/NIA NIH HHS/United States
- R01 MH059587/MH/NIMH NIH HHS/United States
- R01 MH065562/MH/NIMH NIH HHS/United States
- R01 MH059566/MH/NIMH NIH HHS/United States
- R01 AG078657/AG/NIA NIH HHS/United States
- R01 MH125246/MH/NIMH NIH HHS/United States
- R01 MH059588/MH/NIMH NIH HHS/United States
- U01 MH046318/MH/NIMH NIH HHS/United States
- U24 AG087563/AG/NIA NIH HHS/United States
- R01 MH065707/MH/NIMH NIH HHS/United States
- R01 MH060879/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
