Spastin regulates ER-mitochondrial contact sites and mitochondrial homeostasis
- PMID: 39252960
- PMCID: PMC11382127
- DOI: 10.1016/j.isci.2024.110683
Spastin regulates ER-mitochondrial contact sites and mitochondrial homeostasis
Abstract
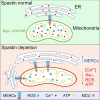
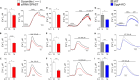
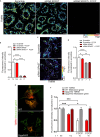
Mitochondria-endoplasmic reticulum (ER) contact sites (MERCs) emerged to play critical roles in numerous cellular processes, and their dysregulation has been associated to neurodegenerative disorders. Mutations in the SPG4 gene coding for spastin are among the main causes of hereditary spastic paraplegia (HSP). Spastin binds and severs microtubules, and the long isoform of this protein, namely M1, spans the outer leaflet of ER membrane where it interacts with other ER-HSP proteins. Here, we showed that overexpressed M1 spastin localizes in ER-mitochondria intersections and that endogenous spastin accumulates in MERCs. We demonstrated in different cellular models that downregulation of spastin enhances the number of MERCs, alters mitochondrial morphology, and impairs ER and mitochondrial calcium homeostasis. These effects are associated with reduced mitochondrial membrane potential, oxygen species levels, and oxidative metabolism. These findings extend our knowledge on the role of spastin in the ER and suggest MERCs deregulation as potential causes of SPG4-HSP disease.
Keywords: Biological sciences; Molecular biology; Molecular interaction.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

