This is a preprint.
The asymmetric opening of HIV-1 Env by a potent CD4 mimetic enables anti-coreceptor binding site antibodies to mediate ADCC
- PMID: 39253431
- PMCID: PMC11383012
- DOI: 10.1101/2024.08.27.609961
The asymmetric opening of HIV-1 Env by a potent CD4 mimetic enables anti-coreceptor binding site antibodies to mediate ADCC
Abstract
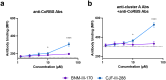
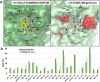
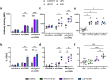
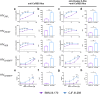
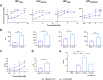
HIV-1 envelope glycoproteins (Env) from primary HIV-1 isolates typically adopt a pretriggered "closed" conformation that resists to CD4-induced (CD4i) non-neutralizing antibodies (nnAbs) mediating antibody-dependent cellular cytotoxicity (ADCC). CD4-mimetic compounds (CD4mcs) "open-up" Env allowing binding of CD4i nnAbs, thereby sensitizing HIV-1-infected cells to ADCC. Two families of CD4i nnAbs, the anti-cluster A and anti-coreceptor binding site (CoRBS) Abs, are required to mediate ADCC in combination with the indane CD4mc BNM-III-170. Recently, new indoline CD4mcs with improved potency and breadth have been described. Here, we show that the lead indoline CD4mc, CJF-III-288, sensitizes HIV-1-infected cells to ADCC mediated by anti-CoRBS Abs alone, contributing to improved ADCC activity. Structural and conformational analyses reveal that CJF-III-288, in combination with anti-CoRBS Abs, potently stabilizes an asymmetric "open" State-3 Env conformation, This Env conformation orients the anti-CoRBS Ab to improve ADCC activity and therapeutic potential.
Figures










References
-
- Finzi D. et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–300 (1997). - PubMed
Publication types
Grants and funding
- T32 AI055403/AI/NIAID NIH HHS/United States
- R01 AI148379/AI/NIAID NIH HHS/United States
- ZIA HD008998/ImNIH/Intramural NIH HHS/United States
- R01 AI150322/AI/NIAID NIH HHS/United States
- F31 AI176650/AI/NIAID NIH HHS/United States
- R01 AI186809/AI/NIAID NIH HHS/United States
- R01 AI174908/AI/NIAID NIH HHS/United States
- P01 AI162242/AI/NIAID NIH HHS/United States
- P01 AI150471/AI/NIAID NIH HHS/United States
- R01 AI176531/AI/NIAID NIH HHS/United States
- R01 AI176904/AI/NIAID NIH HHS/United States
- UM1 AI164562/AI/NIAID NIH HHS/United States
- R37 AI150560/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
