This is a preprint.
Epididymis-specific RNase A family genes regulate fertility and small RNA processing
- PMID: 39253511
- PMCID: PMC11383283
- DOI: 10.1101/2024.08.26.608813
Epididymis-specific RNase A family genes regulate fertility and small RNA processing
Update in
-
Epididymis-specific RNase A family genes regulate fertility and small RNA processing.J Biol Chem. 2024 Dec;300(12):107933. doi: 10.1016/j.jbc.2024.107933. Epub 2024 Oct 28. J Biol Chem. 2024. PMID: 39476961 Free PMC article.
Abstract
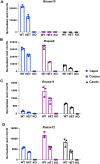
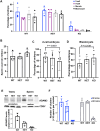
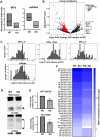
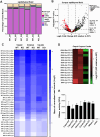
Sperm small RNAs are implicated in intergenerational transmission of paternal environmental effects. Small RNAs generated by cleavage of tRNAs, known as tRNA fragments (tRFs), are an abundant class of RNAs in mature sperm, and can be modulated by environmental conditions. The ribonuclease(s) responsible for the biogenesis of tRFs in the male reproductive tract remains unknown. Angiogenin, a member of the Ribonuclease A superfamily (RNase A), cleaves tRNAs to generate tRFs in response to cellular stress. Four paralogs of Angiogenin, namely Rnase9, Rnase10, Rnase11, and Rnase12, are specifically expressed in the epididymis-a long, convoluted tubule where sperm mature and acquire fertility and motility. The biological functions of these genes remain largely unknown. Here, by generating mice deleted for all four genes (Rnase9-12-/-, termed "KO" for Knock Out), we report that these genes regulate fertility and RNA processing. KO mice showed complete male sterility. KO sperm fertilized oocytes in vitro but failed to efficiently fertilize oocytes in vivo, likely due to an inability of sperm to pass through the utero-tubular junction. Intriguingly, there were decreased levels of fragments of tRNAs (tRFs) and rRNAs (rRNA-derived small RNAs or rsRNAs) in the KO epididymis and epididymal luminal fluid, implying that Rnase9-12 regulate the biogenesis and/or stability of tRFs and rsRNAs. Importantly, KO sperm showed a dramatic decrease in the levels of tRFs, demonstrating a role of Rnase9-12 in regulating sperm RNA composition. Together, our results reveal an unexpected role of four epididymis-specific non-canonical RNase A family genes in fertility and RNA processing.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
