Emergence of dysfunctional neutrophils with a defect in arginase-1 release in severe COVID-19
- PMID: 39253969
- PMCID: PMC11385094
- DOI: 10.1172/jci.insight.171659
Emergence of dysfunctional neutrophils with a defect in arginase-1 release in severe COVID-19
Abstract
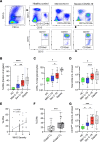
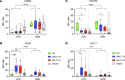
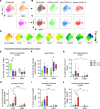
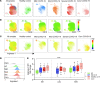
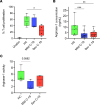
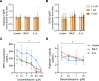
Neutrophilia occurs in patients infected with SARS-CoV-2 (COVID-19) and is predictive of poor outcomes. Here, we link heterogenous neutrophil populations to disease severity in COVID-19. We identified neutrophils with features of cellular aging and immunosuppressive capacity in mild COVID-19 and features of neutrophil immaturity and activation in severe disease. The low-density neutrophil (LDN) number in circulating blood correlated with COVID-19 severity. Many of the divergent neutrophil phenotypes in COVID-19 were overrepresented in the LDN fraction and were less detectable in normal-density neutrophils. Functionally, neutrophils from patients with severe COVID-19 displayed defects in neutrophil extracellular trap formation and reactive oxygen species production. Soluble factors secreted by neutrophils from these patients inhibited T cell proliferation. Neutrophils from patients with severe COVID-19 had increased expression of arginase-1 protein, a feature that was retained in convalescent patients. Despite this increase in intracellular expression, there was a reduction in arginase-1 release by neutrophils into serum and culture supernatants. Furthermore, neutrophil-mediated T cell suppression was independent of arginase-1. Our results indicate the presence of dysfunctional, activated, and immature neutrophils in severe COVID-19.
Keywords: COVID-19; Immunology; Neutrophils.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

