Proteomics profiling and machine learning in nusinersen-treated patients with spinal muscular atrophy
- PMID: 39254732
- PMCID: PMC11387582
- DOI: 10.1007/s00018-024-05426-6
Proteomics profiling and machine learning in nusinersen-treated patients with spinal muscular atrophy
Abstract
Aim: The availability of disease-modifying therapies and newborn screening programs for spinal muscular atrophy (SMA) has generated an urgent need for reliable prognostic biomarkers to classify patients according to disease severity. We aim to identify cerebrospinal fluid (CSF) prognostic protein biomarkers in CSF samples of SMA patients collected at baseline (T0), and to describe proteomic profile changes and biological pathways influenced by nusinersen before the sixth nusinersen infusion (T302).
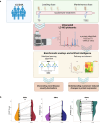
Methods: In this multicenter retrospective longitudinal study, we employed an untargeted liquid chromatography mass spectrometry (LC-MS)-based proteomic approach on CSF samples collected from 61 SMA patients treated with nusinersen (SMA1 n=19, SMA2 n=19, SMA3 n=23) at T0 at T302. The Random Forest (RF) machine learning algorithm and pathway enrichment analysis were applied for analysis.
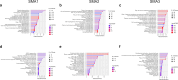
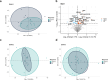
Results: The RF algorithm, applied to the protein expression profile of naïve patients, revealed several proteins that could classify the different types of SMA according to their differential abundance at T0. Analysis of changes in proteomic profiles identified a total of 147 differentially expressed proteins after nusinersen treatment in SMA1, 135 in SMA2, and 289 in SMA3. Overall, nusinersen-induced changes on proteomic profile were consistent with i) common effects observed in allSMA types (i.e. regulation of axonogenesis), and ii) disease severity-specific changes, namely regulation of glucose metabolism in SMA1, of coagulation processes in SMA2, and of complement cascade in SMA3.
Conclusions: This untargeted LC-MS proteomic profiling in the CSF of SMA patients revealed differences in protein expression in naïve patients and showed nusinersen-related modulation in several biological processes after 10 months of treatment. Further confirmatory studies are needed to validate these results in larger number of patients and over abroader timeframe.
Keywords: Artificial intelligence; Biomarkers; Cerebrospinal fluid; Machine learning; Proteomics; Spinal muscular atrophy.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures


References
-
- Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B (2002) Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 70:358–368. 10.1086/338627 10.1086/338627 - DOI - PMC - PubMed
-
- Calucho M, Bernal S, Alías L, March F, Venceslá A, Rodríguez-Álvarez FJ, Aller E, Fernández RM, Borrego S, Millán JM, Hernández-Chico C, Cuscó I, Fuentes-Prior P, Tizzano EF (2018) Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord 28:208–215. 10.1016/j.nmd.2018.01.003 10.1016/j.nmd.2018.01.003 - DOI - PubMed
-
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM (2016) Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388:3017–3026. 10.1016/S0140-6736(16)31408-8 10.1016/S0140-6736(16)31408-8 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

