Single-cell analysis identifies distinct macrophage phenotypes associated with prodisease and proresolving functions in the endometriotic niche
- PMID: 39255000
- PMCID: PMC11420174
- DOI: 10.1073/pnas.2405474121
Single-cell analysis identifies distinct macrophage phenotypes associated with prodisease and proresolving functions in the endometriotic niche
Abstract
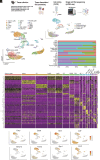
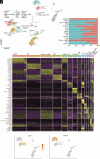
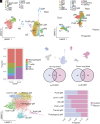
Endometriosis negatively impacts the health-related quality of life of 190 million women worldwide. Novel advances in nonhormonal treatments for this debilitating condition are desperately needed. Macrophages play a vital role in the pathophysiology of endometriosis and represent a promising therapeutic target. In the current study, we revealed the full transcriptomic complexity of endometriosis-associated macrophage subpopulations using single-cell analyses in a preclinical mouse model of experimental endometriosis. We have identified two key lesion-resident populations that resemble i) tumor-associated macrophages (characterized by expression of Folr2, Mrc1, Gas6, and Ccl8+) that promoted expression of Col1a1 and Tgfb1 in human endometrial stromal cells and increased angiogenic meshes in human umbilical vein endothelial cells, and ii) scar-associated macrophages (Mmp12, Cd9, Spp1, Trem2+) that exhibited a phenotype associated with fibrosis and matrix remodeling. We also described a population of proresolving large peritoneal macrophages that align with a lipid-associated macrophage phenotype (Apoe, Saa3, Pid1) concomitant with altered lipid metabolism and cholesterol efflux. Gain of function experiments using an Apoe mimetic resulted in decreased lesion size and fibrosis, and modification of peritoneal macrophage populations in the preclinical model. Using cross-species analysis of mouse and human single-cell datasets, we determined the concordance of peritoneal and lesion-resident macrophage subpopulations, identifying key similarities and differences in transcriptomic phenotypes. Ultimately, we envisage that these findings will inform the design and use of specific macrophage-targeted therapies and open broad avenues for the treatment of endometriosis.
Keywords: endometriosis; heterogeneity; lesion; macrophage; phenotype.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Blériot C., Chakarov S., Ginhoux F., Determinants of resident tissue macrophage identity and function. Immunity 52, 957–970 (2020). - PubMed
-
- Ginhoux F., Jung S., Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014). - PubMed
-
- Ginhoux F., Schultze J. L., Murray P. J., Ochando J., Biswas S. K., New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 17, 34–40 (2016). - PubMed
-
- Schulz C., et al. , A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

