Longitudinal analysis of viral dynamics in HIV+-to-HIV+ HOPE Act kidney-transplant recipients
- PMID: 39255037
- PMCID: PMC11473162
- DOI: 10.1172/JCI181560
Longitudinal analysis of viral dynamics in HIV+-to-HIV+ HOPE Act kidney-transplant recipients
Abstract
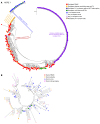
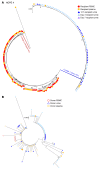
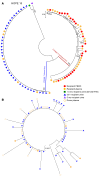
BACKGROUNDThe HIV Organ Policy Equity (HOPE) Act allows individuals living with HIV to accept organs from donors with HIV. This practice widens the pool of available organs, but also presents important virological issues, including the potential for HIV superinfection of the recipient, viral persistence in the kidney, and loss of virological control.METHODSWe addressed these issues by performing in-depth longitudinal viral sequence analyses on urine, blood, and urine-derived renal epithelial cells from 12 recipients of HIV+ kidney allografts.RESULTSWe amplified donor-derived HIV-1 env sequences in 5 out of 12 recipients after transplant. These donor-derived env sequences were amplified from recipient urine, urine-derived renal epithelial cells, and plasma between 12 and 96 hours after transplant and remained detectable up to 16 days after transplant. Env sequences were also detected in kidney biopsies taken from the allografts before implantation in 6 out of the 12 transplant cases, indicating the presence of donor virus within the organ. One recipient had a viremic episode 3.5 years after transplantation as a result of antiretroviral therapy (ART) interruption. Only recipient strain viral sequences were detected in blood, suggesting that the donor virus, if still present, was not reactivated during the temporary ART withdrawal.CONCLUSIONSThis study demonstrates that the HIV env sequences in a donor kidney can be amplified from biopsies taken from the allograft before implantation and can be detected transiently in blood and urine samples collected from the organ recipients after transplantation.FUNDINGNational Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant number R01DK131497.
Keywords: AIDS/HIV; Chronic kidney disease; Organ transplantation; Transplantation.
Figures


Comment in
- HOPE springs eternal: lack of HIV superinfection in HIV Organ Policy Equity Act kidney transplants
Similar articles
-
HOPE springs eternal: lack of HIV superinfection in HIV Organ Policy Equity Act kidney transplants.J Clin Invest. 2024 Oct 15;134(20):e184326. doi: 10.1172/JCI184326. J Clin Invest. 2024. PMID: 39403922 Free PMC article.
-
Outcomes of donor-derived superinfection screening in HIV-positive to HIV-positive kidney and liver transplantation: a multicentre, prospective, observational study.Lancet HIV. 2020 Sep;7(9):e611-e619. doi: 10.1016/S2352-3018(20)30200-9. Epub 2020 Jul 27. Lancet HIV. 2020. PMID: 32730756 Free PMC article.
-
HIV-Positive Kidney Donor Selection for HIV-Positive Transplant Recipients.J Am Soc Nephrol. 2018 Apr;29(4):1090-1095. doi: 10.1681/ASN.2017080853. Epub 2018 Jan 12. J Am Soc Nephrol. 2018. PMID: 29330339 Free PMC article. Review.
-
Utilizing increased risk for disease transmission (IRD) kidneys for pediatric renal transplant recipients.Pediatr Nephrol. 2019 Oct;34(10):1743-1751. doi: 10.1007/s00467-019-04276-w. Epub 2019 Jun 26. Pediatr Nephrol. 2019. PMID: 31243535
-
Increasing the Donor Pool: Organ Transplantation from Donors with HIV to Recipients with HIV.Annu Rev Med. 2021 Jan 27;72:107-118. doi: 10.1146/annurev-med-060419-122327. Annu Rev Med. 2021. PMID: 33502896 Review.
Cited by
-
Inflammation among kidney transplant donors with and without HIV: Multicenter HOPE in Action Consortium.Clin Immunol. 2025 Jul 11;280:110563. doi: 10.1016/j.clim.2025.110563. Online ahead of print. Clin Immunol. 2025. PMID: 40653253
-
HOPE springs eternal: lack of HIV superinfection in HIV Organ Policy Equity Act kidney transplants.J Clin Invest. 2024 Oct 15;134(20):e184326. doi: 10.1172/JCI184326. J Clin Invest. 2024. PMID: 39403922 Free PMC article.
-
Dare to HOPE: a step closer to HIV+-to-HIV+ kidney transplantation as standard of care.Nat Rev Nephrol. 2025 Apr;21(4):220-221. doi: 10.1038/s41581-024-00920-3. Nat Rev Nephrol. 2025. PMID: 39695335 No abstract available.
References
-
- Arnold E. The HIV organ policy equity act: offering hope to individuals with end stage renal disease and HIV. Nephrol Nurs J. 2017;44(3):230–249. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

