PDE4B Missense Variant Increases Susceptibility to Post-traumatic Stress Disorder-Relevant Phenotypes in Mice
- PMID: 39256048
- PMCID: PMC11502227
- DOI: 10.1523/JNEUROSCI.0137-24.2024
PDE4B Missense Variant Increases Susceptibility to Post-traumatic Stress Disorder-Relevant Phenotypes in Mice
Abstract
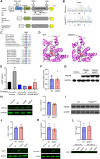
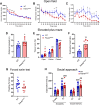
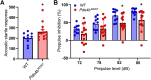
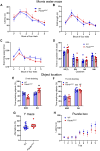
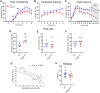
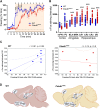
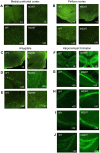
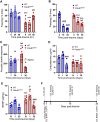
Large-scale genome-wide association studies (GWASs) have associated intronic variants in PDE4B, encoding cAMP-specific phosphodiesterase-4B (PDE4B), with increased risk for post-traumatic stress disorder (PTSD), as well as schizophrenia and substance use disorders that are often comorbid with it. However, the pathophysiological mechanisms of genetic risk involving PDE4B are poorly understood. To examine the effects of PDE4B variation on phenotypes with translational relevance to psychiatric disorders, we focused on PDE4B missense variant M220T, which is present in the human genome as rare coding variant rs775201287. When expressed in HEK-293 cells, PDE4B1-M220T exhibited an attenuated response to a forskolin-elicited increase in the intracellular cAMP concentration. In behavioral tests, homozygous Pde4b M220T male mice with a C57BL/6JJcl background exhibited increased reactivity to novel environments, startle hyperreactivity, prepulse inhibition deficits, altered cued fear conditioning, and enhanced spatial memory, accompanied by an increase in cAMP signaling pathway-regulated expression of BDNF in the hippocampus. In response to a traumatic event (10 tone-shock pairings), neuronal activity was decreased in the cortex but enhanced in the amygdala and hippocampus of Pde4b M220T mice. At 24 h post-trauma, Pde4b M220T mice exhibited increased startle hyperreactivity and decreased plasma corticosterone levels, similar to phenotypes exhibited by PTSD patients. Trauma-exposed Pde4b M220T mice also exhibited a slower decay in freezing at 15 and 30 d post-trauma, demonstrating enhanced persistence of traumatic memories, similar to that exhibited by PTSD patients. These findings provide substantive mouse model evidence linking PDE4B variation to PTSD-relevant phenotypes and thus highlight how genetic variation of PDE4B may contribute to PTSD risk.
Keywords: PDE4B; PTSD; cAMP; fear memory; schizophrenia; trauma.
Copyright © 2024 Lipina et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- American Psychiatric Association (2013) Diagnostic and statistical manual of mental disorders, Ed 5. Arlington, VA: American Psychiatric Association.
-
- Armstrong P, Güngör H, Anongjanya P, Tweedy C, Parkin E, Johnston J, Carr IM, Dawson N, Clapcote SJ (2024) Protective effect of PDE4B subtype-specific inhibition in an App knock-in mouse model for Alzheimer's disease. Neuropsychopharmacology 49:1559–1568. 10.1038/s41386-024-01852-z - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
