Endurance exercise-induced histone methylation modification involved in skeletal muscle fiber type transition and mitochondrial biogenesis
- PMID: 39256490
- PMCID: PMC11387812
- DOI: 10.1038/s41598-024-72088-6
Endurance exercise-induced histone methylation modification involved in skeletal muscle fiber type transition and mitochondrial biogenesis
Abstract
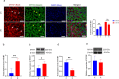
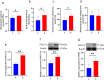
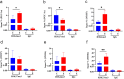
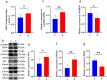
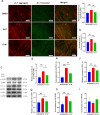
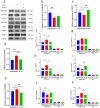
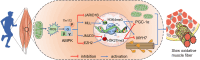
Skeletal muscle is a highly heterogeneous tissue, and its contractile proteins are composed of different isoforms, forming various types of muscle fiber, each of which has its own metabolic characteristics. It has been demonstrated that endurance exercise induces the transition of muscle fibers from fast-twitch to slow-twitch muscle fiber type. Herein, we discover a novel epigenetic mechanism for muscle contractile property tightly coupled to its metabolic capacity during muscle fiber type transition with exercise training. Our results show that an 8-week endurance exercise induces histone methylation remodeling of PGC-1α and myosin heavy chain (MHC) isoforms in the rat gastrocnemius muscle, accompanied by increased mitochondrial biogenesis and an elevated ratio of slow-twitch to fast-twitch fibers. Furthermore, to verify the roles of reactive oxygen species (ROS) and AMPK in exercise-regulated epigenetic modifications and muscle fiber type transitions, mouse C2C12 myotubes were used. It was shown that rotenone activates ROS/AMPK pathway and histone methylation enzymes, which then promote mitochondrial biogenesis and MHC slow isoform expression. Mitoquinone (MitoQ) partially blocking rotenone-treated model confirms the role of ROS in coupling mitochondrial biogenesis with muscle fiber type. In conclusion, endurance exercise couples mitochondrial biogenesis with MHC slow isoform by remodeling histone methylation, which in turn promotes the transition of fast-twitch to slow-twitch muscle fibers. The ROS/AMPK pathway may be involved in the regulation of histone methylation enzymes by endurance exercise.
Keywords: AMPK; Endurance exercise; Histone methylation; Mitochondrial biogenesis; ROS; Skeletal muscle fiber type.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

