Establishment of single-cell transcriptional states during seed germination
- PMID: 39256563
- PMCID: PMC11410669
- DOI: 10.1038/s41477-024-01771-3
Establishment of single-cell transcriptional states during seed germination
Abstract
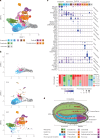
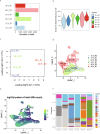
Germination involves highly dynamic transcriptional programs as the cells of seeds reactivate and express the functions necessary for establishment in the environment. Individual cell types have distinct roles within the embryo, so must therefore have cell type-specific gene expression and gene regulatory networks. We can better understand how the functions of different cell types are established and contribute to the embryo by determining how cell type-specific transcription begins and changes through germination. Here we describe a temporal analysis of the germinating Arabidopsis thaliana embryo at single-cell resolution. We define the highly dynamic cell type-specific patterns of gene expression and how these relate to changing cellular function as germination progresses. Underlying these are unique gene regulatory networks and transcription factor activity. We unexpectedly discover that most embryo cells transition through the same initial transcriptional state early in germination, even though cell identity has already been established during embryogenesis. Cells later transition to cell type-specific gene expression patterns. Furthermore, our analyses support previous findings that the earliest events leading to the induction of seed germination take place in the vasculature. Overall, our study constitutes a general framework with which to characterize Arabidopsis cell transcriptional states through seed germination, allowing investigation of different genotypes and other plant species whose seed strategies may differ.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














Comment in
-
Understanding cell-type-specific regulation during seed germination.Nat Plants. 2024 Sep;10(9):1295-1296. doi: 10.1038/s41477-024-01772-2. Nat Plants. 2024. PMID: 39256565 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

