The evolutionary theory of cancer: challenges and potential solutions
- PMID: 39256635
- PMCID: PMC11627115
- DOI: 10.1038/s41568-024-00734-2
The evolutionary theory of cancer: challenges and potential solutions
Abstract
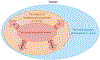
The clonal evolution model of cancer was developed in the 1950s-1970s and became central to cancer biology in the twenty-first century, largely through studies of cancer genetics. Although it has proven its worth, its structure has been challenged by observations of phenotypic plasticity, non-genetic forms of inheritance, non-genetic determinants of clone fitness and non-tree-like transmission of genes. There is even confusion about the definition of a clone, which we aim to resolve. The performance and value of the clonal evolution model depends on the empirical extent to which evolutionary processes are involved in cancer, and on its theoretical ability to account for those evolutionary processes. Here, we identify limits in the theoretical performance of the clonal evolution model and provide solutions to overcome those limits. Although we do not claim that clonal evolution can explain everything about cancer, we show how many of the complexities that have been identified in the dynamics of cancer can be integrated into the model to improve our current understanding of cancer.
© 2024. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

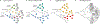


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

