Exosome-mediated transfer of lncRNA RP3-340B19.3 promotes the progression of breast cancer by sponging miR-4510/MORC4 axis
- PMID: 39256868
- PMCID: PMC11389435
- DOI: 10.1186/s12935-024-03490-3
Exosome-mediated transfer of lncRNA RP3-340B19.3 promotes the progression of breast cancer by sponging miR-4510/MORC4 axis
Abstract
Background: This study aims to explore the molecular mechanism of lncRNA RP3-340B19.3 on breast cancer cell proliferation and metastasis and clinical significance of lncRNA RP3-340B19.3 for breast cancer.
Methods: The subcellular localization of lncRNA RP3-340B19.3 was identified using RNA fluorescence in situ hybridization (FISH). The expression of lncRNA RP3-340B19.3 in breast cancer cells, breast cancer tissues, as well as the serum and serum exosomes of breast cancer patients, was measured through quantitative RT-PCR. In the in vitro setting, we conducted experiments to observe the effects of RP3-340B19.3 on both cell migration and proliferation. This was achieved through the utilization of transwell migration assays as well as clone formation assays. Meanwhile, transwell migration assays and clone formation assays were used to observe the effects of MDA-MB-231-exosomes enriched in RP3-340B19.3 on breast cancer microenvironment cells MCF7 and BMMSCs. Additionally, western blotting techniques were used to assess the expression levels of proteins associated with essential cellular processes such as proliferation, apoptosis, and metastasis. In vivo, the impact of RP3-340B19.3 knockdown on tumour weight and volume was observed within a nude mice model. We aimed to delve into the intricate molecular mechanisms involving RP3-340B19.3 by using bioinformatics analysis, dual luciferase reporter gene experiments and western blotting. Moreover, the potential correlations between RP3-340B19.3 expression and various clinical pathological characteristics were analyzed.
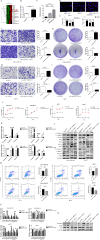
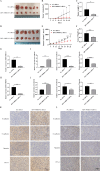
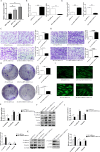
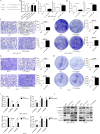
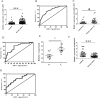
Results: Our investigation revealed that RP3-340B19.3 was expressed in both the cytoplasm and nucleus, with a noteworthy increase in breast cancer cells. Notably, we found that RP3-340B19.3 exerted a promoting influence on the proliferation and migration of breast cancer cells, both in vitro and in vivo. MDA-MB-231-exosomes enriched in RP3-340B19.3 promoted the proliferation and migration of MCF7 and BMMSCs in vitro. Mechanistically, RP3-340B19.3 demonstrated the capability to modulate the expression of MORC4 by forming a complex with miR-4510. This interaction subsequently triggered the activation of the NF-κB and Wnt-β-catenin signaling pathways. Furthermore, our study highlighted the potential diagnostic utility of RP3-340B19.3. We discovered its presence in the serum and exosomes of breast cancer patients, showing promising efficacy as a diagnostic marker. Notably, the diagnostic potential of RP3-340B19.3 was particularly significant in relation to distinguishing between different pathological types of breast cancer and correlating with tumour diameter.
Conclusion: Our findings establish that RP3-340B19.3 plays a pivotal role in driving the proliferation and metastasis of breast cancer. Additionally, exosomes enriched in RP3-340B19.3 could influence MCF7 and BMMSCs in tumour microenvironment, promoting the progression of breast cancer. This discovery positions RP3-340B19.3 as a prospective novel candidate for a tumour marker, offering substantial potential in the realms of breast cancer diagnosis and treatment strategies.
Keywords: Breast cancer; Exosome; Metastasis; Proliferation; lncRNA RP3-340B19.3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Grants and funding
- no.SKJY2021097/Science and Technology Program of Suzhou
- no.SKY2021045/Science and Technology Program of Suzhou
- no. SH2022044/The Key Research and Development Project of Zhenjiang
- no.KJXW2020019/Suzhou Science, Education and Health Youth Science and Technology Project
- no.81702078/National Natural Science Foundation of China
- no.BK20170356/the Natural Science Foundation of Jiangsu Province
- no.XKTJ-HRC2021001/Gusu Talent Program; Support for the project of nuclear technology medical application supported by discipline construction
- no.FYX202123/Maternal and Child Health Association Project of Jiangsu province
- Qngg2023008/"National Tutor System" Training Program for Health Youth Key Talents in Suzhou
LinkOut - more resources
Full Text Sources
Miscellaneous

