SlDELLA interacts with SlPIF4 to regulate arbuscular mycorrhizal symbiosis and phosphate uptake in tomato
- PMID: 39257536
- PMCID: PMC11384114
- DOI: 10.1093/hr/uhae195
SlDELLA interacts with SlPIF4 to regulate arbuscular mycorrhizal symbiosis and phosphate uptake in tomato
Abstract
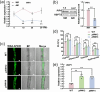
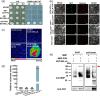
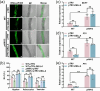
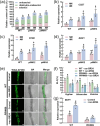
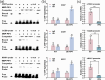
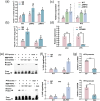
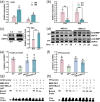
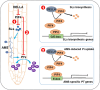
Arbuscular mycorrhizal symbiosis (AMS), a complex and delicate process, is precisely regulated by a multitude of transcription factors. PHYTOCHROME-INTERACTING FACTORS (PIFs) are critical in plant growth and stress responses. However, the involvement of PIFs in AMS and the molecular mechanisms underlying their regulator functions have not been well elucidated. Here, we show that SlPIF4 negatively regulates the arbuscular mycorrhizal fungi (AMF) colonization and AMS-induced phosphate uptake in tomato. Protein-protein interaction studies suggest that SlDELLA interacts with SlPIF4, reducing its protein stability and inhibiting its transcriptional activity towards downstream target genes. This interaction promotes the accumulation of strigolactones (SLs), facilitating AMS development and phosphate uptake. As a transcription factor, SlPIF4 directly transcriptionally regulates genes involved in SLs biosynthesis, including SlCCD7, SlCDD8, and SlMAX1, as well as the AMS-specific phosphate transporter genes PT4 and PT5. Collectively, our findings uncover a molecular mechanism by which the SlDELLA-SlPIF4 module regulates AMS and phosphate uptake in tomato. We clarify a molecular basis for how SlPIF4 interacts with SLs to regulate the AMS and propose a potential strategy to improve phosphate utilization efficiency by targeting the AMS-specific phosphate transporter genes PTs.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
No conflicting interest.
Figures
References
-
- Johnston AE, Poulton PR, Fixen PE. et al. Phosphorus: its efficient use in agriculture. Adv Agron. 2014;123:177–228
-
- Oldroyd GED, Leyser O. A plant's diet, surviving in a variable nutrient environment. Science. 2020;368:eaba0196. - PubMed
-
- Genre A, Lanfranco L, Perotto S. et al. Unique and common traits in mycorrhizal symbioses. Nat Rev Microbiol. 2020;18:649–60 - PubMed
-
- Shi JC, Wang XL, Wang E. Mycorrhizal symbiosis in plant growth and stress adaptation: from genes to ecosystems. Annu Rev Plant Biol. 2023;74:569–607 - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

