Lactylation signature identifies liver fibrosis phenotypes and traces fibrotic progression to hepatocellular carcinoma
- PMID: 39257588
- PMCID: PMC11383765
- DOI: 10.3389/fimmu.2024.1433393
Lactylation signature identifies liver fibrosis phenotypes and traces fibrotic progression to hepatocellular carcinoma
Abstract
Introduction: Precise staging and classification of liver fibrosis are crucial for the hierarchy management of patients. The roles of lactylation are newly found in the progression of liver fibrosis. This study is committed to investigating the signature genes with histone lactylation and their connection with immune infiltration among liver fibrosis with different phenotypes.
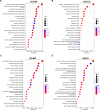
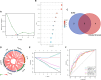
Methods: Firstly, a total of 629 upregulated and 261 downregulated genes were screened out of 3 datasets of patients with liver fibrosis from the GEO database and functional analysis confirmed that these differentially expressed genes (DEGs) participated profoundly in fibrosis-related processes. After intersecting with previously reported lactylation-related genes, 12 DEGs related to histone lactylation were found and narrowed down to 6 core genes using R algorithms, namely S100A6, HMGN4, IFI16, LDHB, S100A4, and VIM. The core DEGs were incorporated into the Least absolute shrinkage and selection operator (LASSO) model to test their power to distinguish the fibrotic stage.
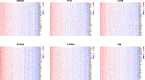
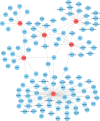
Results: Advanced fibrosis presented a pattern of immune infiltration different from mild fibrosis, and the core DEGs were significantly correlated with immunocytes. Gene set and enrichment analysis (GSEA) results revealed that core DEGs were closely linked to immune response and chemokine signaling. Samples were classified into 3 clusters using the LASSO model, followed by gene set variation analysis (GSVA), which indicated that liver fibrosis can be divided into status featuring lipid metabolism reprogramming, immunity immersing, and intermediate of both. The regulatory networks of the core genes shared several transcription factors, and certain core DEGs also presented dysregulation in other liver fibrosis and idiopathic pulmonary fibrosis (IPF) cohorts, indicating that lactylation may exert comparable functions in various fibrotic pathology. Lastly, core DEGs also exhibited upregulation in HCC.
Discussion: Lactylation extensively participates in the pathological progression and immune infiltration of fibrosis. Lactylation and related immune infiltration could be a worthy focus for the investigation of HCC developed from liver fibrosis.
Keywords: hepatocellular carcinoma; immune infiltration; lactylation; liver fibrosis; machine learning.
Copyright © 2024 Li, Li, Xiao and Lai.
Conflict of interest statement
Author W-wL was employed by the company Guangzhou Wondfo Health Science and Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

