This is a preprint.
High-throughput synthesis and specificity characterization of natively paired antibodies using oPool+ display
- PMID: 39257766
- PMCID: PMC11383711
- DOI: 10.1101/2024.08.30.610421
High-throughput synthesis and specificity characterization of natively paired antibodies using oPool+ display
Update in
-
High-throughput synthesis and specificity characterization of natively paired influenza hemagglutinin antibodies with oPool+ display.Sci Transl Med. 2025 Sep 3;17(814):eadt4147. doi: 10.1126/scitranslmed.adt4147. Epub 2025 Sep 3. Sci Transl Med. 2025. PMID: 40901925 Free PMC article.
Abstract
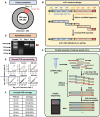
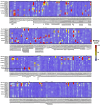
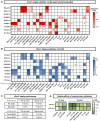
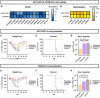
Antibody discovery is crucial for developing therapeutics and vaccines as well as understanding adaptive immunity. However, the lack of approaches to synthesize antibodies with defined sequences in a high-throughput manner represents a major bottleneck in antibody discovery. Here, we presented oPool+ display, a high-throughput cell-free platform that combined oligo pool synthesis and mRNA display to rapidly construct and characterize many natively paired antibodies in parallel. As a proof-of-concept, we applied oPool+ display to probe the binding specificity of >300 uncommon influenza hemagglutinin (HA) antibodies against 9 HA variants through 16 different screens. Over 5,000 binding tests were performed in 3-5 days with further scaling potential. Follow-up structural analysis of two HA stem antibodies revealed the previously unknown versatility of IGHD3-3 gene segment in recognizing the HA stem. Overall, this study established an experimental platform that not only accelerate antibody characterization, but also enable unbiased discovery of antibody molecular signatures.
Keywords: B-cell receptors; High-throughput screening; antibody; broadly neutralizing; cryo-EM; hemagglutinin; influenza; mRNA display; oligo pools.
Conflict of interest statement
DECLARATION OF INTERESTS N.C.W. consults for HeliXon. The authors declare no other competing interests.
Figures


References
-
- Sheets M.D., Amersdorfer P., Finnern R., Sargent P., Lindquist E., Schier R., Hemingsen G., Wong C., Gerhart J.C., and Marks J.D. (1998). Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc. Natl. Acad. Sci. U. S. A. 95, 6157–6162. 10.1073/pnas.95.11.6157. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
