This is a preprint.
O-glycosylation contributes to mammalian glycoRNA biogenesis
- PMID: 39257776
- PMCID: PMC11384000
- DOI: 10.1101/2024.08.28.610074
O-glycosylation contributes to mammalian glycoRNA biogenesis
Abstract
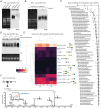
There is an increasing appreciation for the role of cell surface glycans in modulating interactions with extracellular ligands and participating in intercellular communication. We recently reported the existence of sialoglycoRNAs, where mammalian small RNAs are covalently linked to N-glycans through the modified base acp3U and trafficked to the cell surface. However, little is currently known about the role for O-glycosylation, another major class of carbohydrate polymer modifications. Here, we use parallel genetic, enzymatic, and mass spectrometry approaches to demonstrate that O-linked glycan biosynthesis is responsible for the majority of sialoglycoRNA levels. By examining the O-glycans associated with RNA from cell lines and colon organoids we find known and previously unreported O-linked glycan structures. Further, we find that O-linked glycans released from small RNA from organoids derived from ulcerative colitis patients exhibit higher levels of sialylation than glycans from healthy organoids. Together, our work provides flexible tools to interrogate O-linked glycoRNAs (O-glycoRNA) and suggests that they may be modulated in human disease.
Conflict of interest statement
DECLARATION OF INTERESTS R.A.F. is a stockholder of ORNA Therapeutics. R.A.F. is a board of directors member and stockholder of Chronus Health and Blue Planet Systems. The other authors declare no competing interests.
Figures


References
-
- Flynn R.A., Pedram K., Malaker S.A., Batista P.J., Smith B.A.H., Johnson A.G., George B.M., Majzoub K., Villalta P.W., Carette J.E., et al. (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124.e22. 10.1016/j.cell.2021.04.023. - DOI - PMC - PubMed
-
- Bi M., Zhang Z., Wang T., Liang H., and Tian Z. (2023). A draft of human N-glycans of glycoRNA. bioRxiv, 2023.09.18.558371. 10.1101/2023.09.18.558371. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
