This is a preprint.
Self-Assembled Generation of Multi-zonal Liver Organoids from Human Pluripotent Stem Cells
- PMID: 39257824
- PMCID: PMC11384014
- DOI: 10.1101/2024.08.30.610426
Self-Assembled Generation of Multi-zonal Liver Organoids from Human Pluripotent Stem Cells
Update in
-
Multi-zonal liver organoids from human pluripotent stem cells.Nature. 2025 May;641(8065):1258-1267. doi: 10.1038/s41586-025-08850-1. Epub 2025 Apr 16. Nature. 2025. PMID: 40240593 Free PMC article.
Abstract
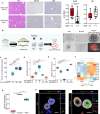
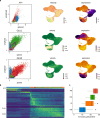
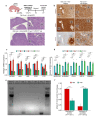
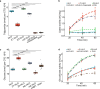
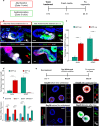
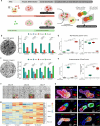
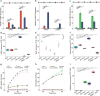
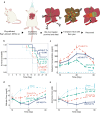
Distinct hepatocyte subpopulations are spatially segregated along the portal-central axis and critical to understanding metabolic homeostasis and liver injury. While several bioactive molecules have been described to play a role in directing zonal fates, including ascorbate and bilirubin, in vitro replication of zonal liver architecture has not been achieved to date. In order to evaluate hepatic zonal polarity, we developed a self-assembling zone-specific liver organoid culture by co-culturing ascorbate and bilirubin enriched hepatic progenitors derived from human induced pluripotent stem cells. We found that preconditioned hepatocyte-like cells exhibited zone-specific functions associated with urea cycle, glutathione synthesis and glutamate synthesis. Single nucleus RNA sequencing analysis of these zonally patterned organoids identifies hepatoblast differentiation trajectory that mimics periportal-, interzonal-, and pericentral human hepatocytes. Epigenetic and transcriptomic analysis showed that zonal identity is orchestrated by ascorbate or bilirubin dependent binding of histone acetyltransferase p300 (EP300) to methylcytosine dioxygenase TET1 or hypoxia-inducible factor 1-alpha (HIF1α). Transplantation of the self-assembled zonally patterned human organoids improved survival of immunodeficient rats who underwent bile duct ligation by ameliorating the hyperammonemia and hyperbilirubinemia. Overall, this multi-zonal organoid system serves as an in vitro human model to better recapitulate hepatic architecture relevant to liver development and disease.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures









References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
